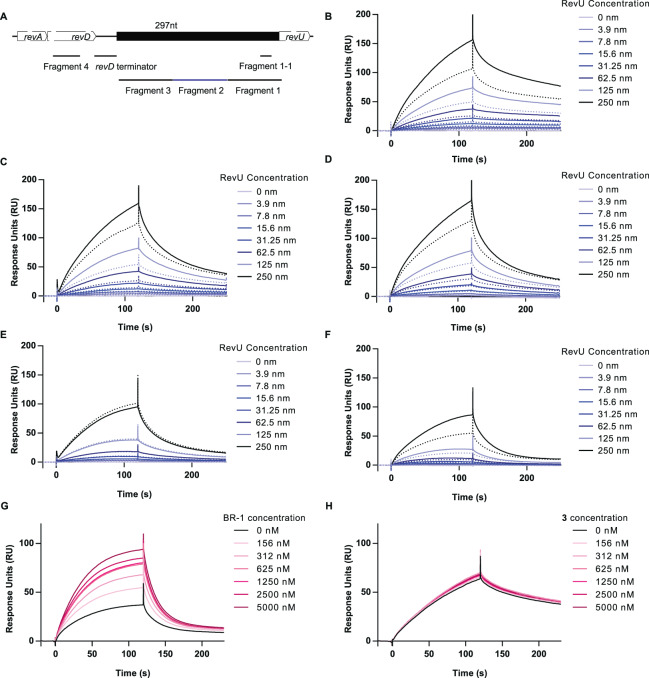
Figure 5.
SPR analysis of the DNA-binding activity of RevU in the presence of BR-1. (A) Schematic representation of the revU promoter region and DNA fragments used for evaluating the RevU promoter–BR-1 interaction. Fragments 1–3, 99 bp in length, are located immediately upstream of the revU start codon. Fragment 4 (50-bp) located on revD coding sequence (GATTATGCGTCGCATTCGGTGTTTGTGGAGTTGATCGAGGATCGGGTTCT) was used for negative control. The underlined sequences were also found in another part of revA and revD sequence). The revD terminator sequence (CACCCAGCCCTCCCGCGGGAGCCGCCCGGCTCCCGCGGAAGGCGCCCGCG) lies upstream of fragment 3. Fragment 1-1 (22 bp: ACGCCGCAACGACCAACAGAGG) contains the putative lux-box sequence. Blank-subtracted SPR sensorgram showing the binding of biotin-labelled DNA fragment 1 (B), fragment 2 (C), fragment 3 (D), fragment 4 (E), and fragment 1-1 (F) to RevU in the presence (solid lines) or absence (dotted lines) of 1.25 µM BR-1. RevU was injected at various concentrations across the chip surface. Blank-subtracted SPR sensorgram showing the binding of biotinylated DNA fragment 1-1 to RevU (125 nM). Various concentration of BR-1 (G) and 3 (H) were tested. Fold changes in response unit BR-1 (+/−) is shown in Fig. S5.