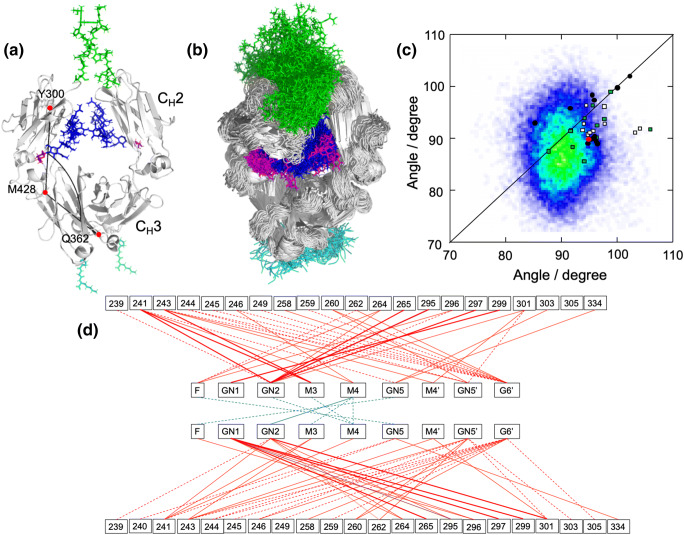
Fig. 2.
Dynamic view of IgG-Fc provided by experimentally validated computational approach. (a) The starting structure of SAXS-validated MD simulation based on the crystal structure of fucosyl IgG1-Fc (3AVE) supplemented with the hinge (green) and C-terminal (cyan) segments along with the terminal galactose residues (magenta) of the Manα1–6 branches. N-glycans are depicted in blue except for the terminal galactose. The intra-chain domain-orientation angle between CH2 and CH3 defined by Cα atoms of Y300, M428, and Q362 are shown in chain A. (b) The superposition of 256 structures extracted every 100 ns from the MD trajectory. The structures were visualized by PyMOL (https://www.pymol.org). (c) Distribution of intra-chain domain-orientation angles between CH2 and CH3 for ensemble models of IgG1-Fc and various crystal structures of IgG1-Fc. Angles between CH2 and CH3 domains of chain A and chain B are plotted on the x and y axes, respectively. Data depict results from ensemble models derived from MD simulations along with angles observed in crystal structures [circles for uncomplexed Fc (red, the starting structures used for the corresponding MD simulations; black, Fc with native N-glycans), rectangles for complexed Fc structures (white, complex with sFcγRs; green, complex with other ligands)]. (d) Pairs of contact residues found within 4 Å in the ensemble model derived from the MD simulation are connected by different types of line segments (red for carbohydrate-protein contact and cyan for carbohydrate-carbohydrate contact) according to incidence as follows: more than 24,000 pairs (thick solid line), 24,000 to 16,000 pairs (thin solid line), and 16,000 to 8000 pairs (dashed line). Adapted from reference (Yanaka et al. 2019a) with modification