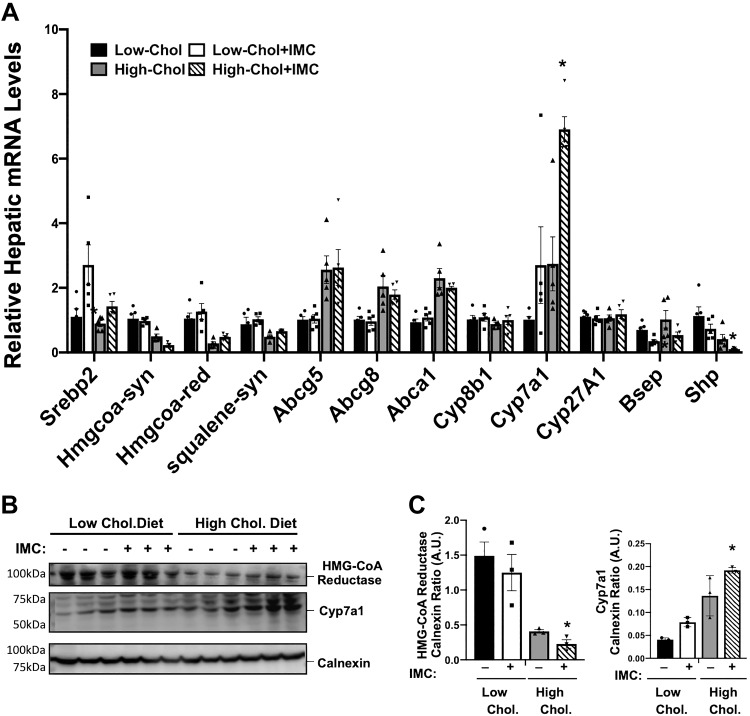
Fig. 4.
Trimethylamine (TMA) lyase inhibition alters the hepatic expression of key genes involved in sterol and bile acid metabolism. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on these diets, liver RNA was extracted and gene expression was measured using quantitative real-time PCR. A: relative mRNA expression of sterol regulatory-binding protein 2 (Srebp2), HMG-CoA reductase (HMG CoA-red), HMG-CoA synthase (HMG CoA-syn), squalene synthase (Squalene-syn), ATP-binding cassette transporters (Abca1, Abcg5, and Abcg8), bile acid modifying cytochrome-P450 family members (Cyp7a1, Cyp8b1, and Cyp27a1), bile salt export pump (Bsep), and small heterodimeric partner (Shp) were quantified using the ΔΔCT method. B and C: Western blot analysis HMG-CoA reductase and Cyp7a1 with densitometric quantification. Data shown in A represent the means ± SE for n = 5 mice per group, whereas data in B and C represent the means ± SE for n = 3 mice per group; *significantly different than the nondrug-treated mice within each diet group. AU, arbitrary units.