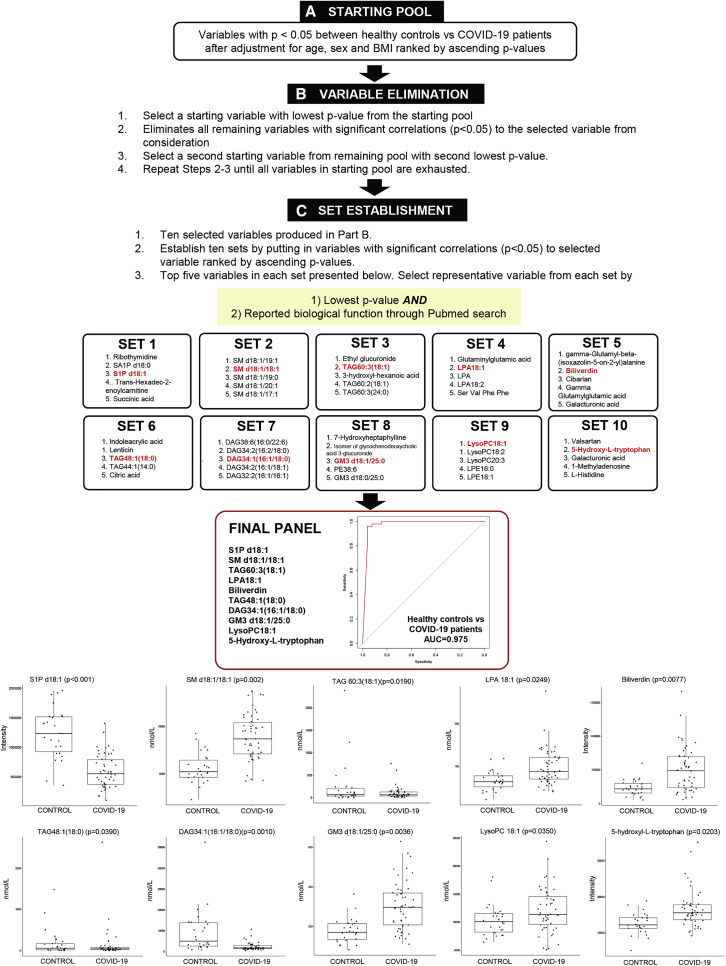
Figure 1.
Plasma Panel for Differentiating COVID-19 Patients from Heathy Controls
Overview of selection scheme for plasma metabolite panel to differentiate COVID-19 (n = 50) patients from healthy controls (n = 26).
(A) From a total of 1,002 variables measured (598 lipids and 404 polar metabolites), variables with p < 0.05 between healthy controls and COVID-19 patients after adjustment for age, sex, and BMI were sieved out to form a starting pool comprising 322 variables.
(B) A starting variable with the lowest p value was selected, and variables with significant correlations (p < 0.05) to the starting variable selected were removed from consideration. From remaining variables in the starting pool, the next starting variable with the second lowest p value was identified, and the process was repeated in an iterative fashion until all variables in the starting pool were exhausted. This process generated a list of ten variables.
(C) Variables with significant correlations (p < 0.05) to each of the selected variables were added together to form ten established sets. To select a representative variable from each established set, the variable with the smallest p value and with reported biological function from a Pubmed search was chosen. A final panel of ten plasma metabolites, including S1P d18:1, SM d18:1/18:1, TAG60:3(18:1), LPA 18:1, biliverdin, TAG 48:1(18:0), DAG34:1(16:1/18:0), GM3 d18:1/25:0, lysoPC18:1, and 5-hydroxy-L-tryptophan, was generated, which distinguished between healthy controls and COVID-19 patients with an area under the curve (AUC) = 0.975 in a logistic regression model with leave-one-out (LOO) cross-validation. Boxplots for the ten selected metabolites in the final panel were illustrated and p values were indicated on top of each boxplot. Levels of polar metabolites measured using untargeted metabolomics were presented as corrected intensities, and lipids quantitated using targeted lipidomics were presented in nanomoles of lipids per liter (nmol/L) plasma.
See also Figures S2–S4.