Figure 6.
Identified S Mutations Are Unlikely to Affect Epitopes Revealed by nsEMPEM and Single-Particle Cryo-EM
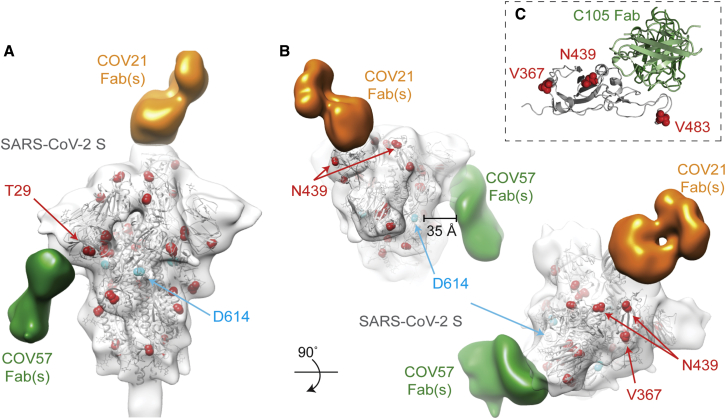
(A and B) Side (A) and top (B) views of refined 3D model of SARS-CoV-2 S trimer alone fitted with a reference structure (PDB: 6VYB; gray cartoon) to illustrate locations of mutations observed in circulating SARS-CoV-2 isolates (Table S3) (red spheres). Residues affected by mutations that are disordered in the SARS-CoV-2 S structure (V483A) or in regions that are not included in the S ectodomain (signal sequence or cytoplasmic tail) are not shown. Densities corresponding to Fabs were separated, colored, and displayed on the same 3D volume.
(C) C105-RBD interaction from the cryo-EM structure of the C105-S complex (Figure 5) showing locations of RBD mutations. V483 is ordered in this structure.