Figure 7.
S Protein Epitopes Offer Different Possibilities for Avidity Effects during IgG and Receptor Binding
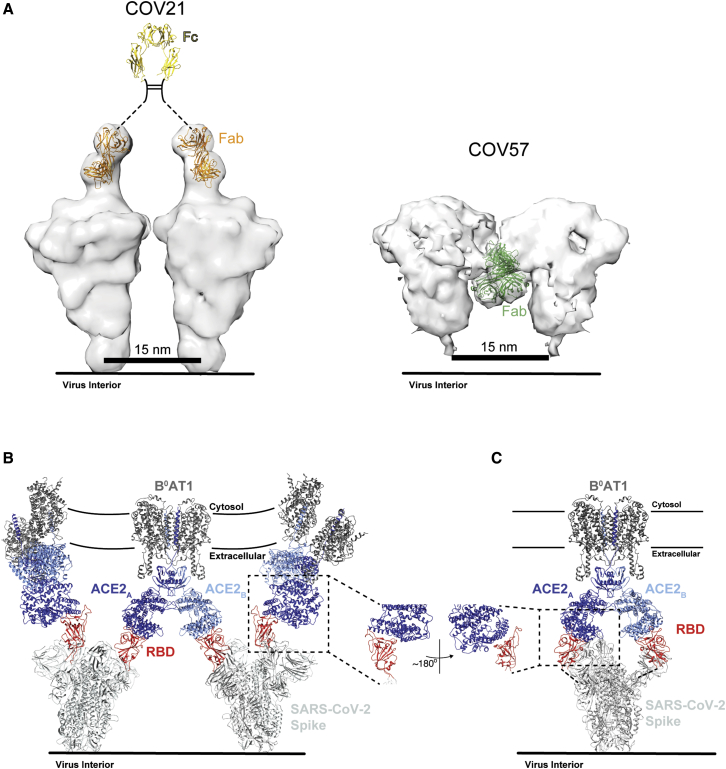
(A) Left: model of two adjacent S trimers separated by ∼15 nm, as seen on coronaviruses by cryo-electron tomography (Neuman et al., 2011), demonstrating that the orientation of COV21 Fab(s) on S could accommodate inter-spike crosslinking by a single IgG. The Fc portion of the IgG (PDB: 1IGT) was modeled assuming flexibility between the Fabs and the Fc (Sandin et al., 2004) and with the hinge region indicated by a dotted line since it is disordered in crystal structures of intact IgGs (Harris et al., 1992; Harris et al., 1998; Saphire et al., 2001). Right: Example of a model of two adjacent S trimers with bound Fab(s) in the orientation observed in the COV57 Fab(s)-S reconstruction demonstrating that inter-spike crosslinking is unlikely due to the “downward” orientation of the Fab(s), which does not permit linking by an Fc region, and predicted steric clashes between adjacent Fabs. Inter-spike crosslinking is also not possible for other orientations of two adjacent COV57 Fab(s)-S complexes (not shown).
(B) Model of S trimers with two RBDs in an “up” position based on a cryo-EM structure of SARS-CoV S trimer (Kirchdoerfer et al., 2018) (PDB: 6CRX) interacting with full-length ACE2 receptors from the cryo-EM structure of soluble SARS-CoV-2 RBDs bound to the dimeric membrane form of ACE2 (Yan et al., 2020) (PDB: 6M17). Inter-spike crosslinking is possible if ACE2 dimers cluster in the membrane.
(C) Model of intra-spike crosslinking between dimeric ACE2 and an S protein trimer with two RBDs in an “up” position. The RBDs were rotated by ∼180° about their long axes to allow binding of the ACE2 ectodomains. Rotation of the RBD is a possibility since its position is flexible with respect to the remaining part of the S trimer (Walls et al., 2019). In this model, RBDs from a single S trimer could bind the ACE2 dimer in the same configuration as seen in the BoAT1-ACE2-SARS-CoV-2 RBD structure (PDB: 6M17).