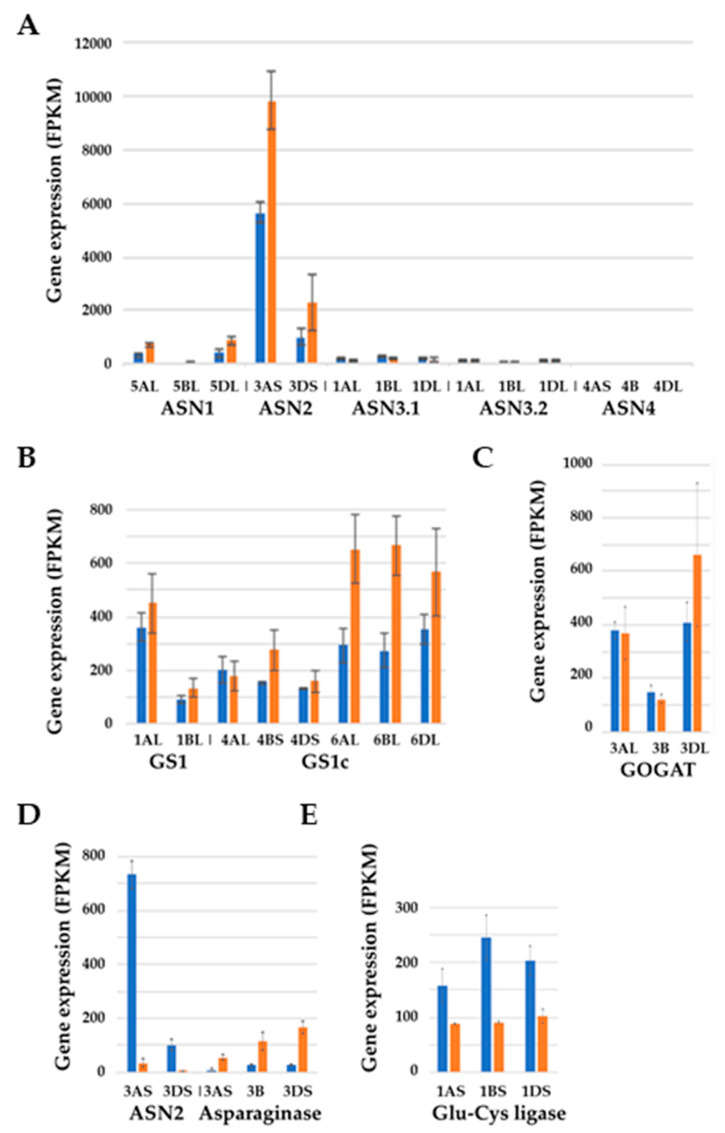
Figure 5.
Graphical representation (means and standard errors) showing the effects of sulphur on the expression levels (fragments per kilobase of transcript per million mapped reads (FPKM)) of selected genes in developing grain of wheat (Triticum aestivum) genotype SR3 at 21 days post-anthesis. Sulphur was either supplied (blue columns) or withheld (brown columns). Results for each homeologue are shown separately, with chromosomal locations indicated. (A) Expression of asparagine synthetase genes ASN1, ASN2, ASN3.1, ASN3.2, and ASN4 in the embryo. The increase in expression of the ASN1 homeologue on chromosome 5D in response to sulphur deficiency was significant (p = 0.0478), as was the increase in expression of both ASN2 homeologues (p = 0.038 and 0.047 for the 3A and 3D homeologues, respectively). (B) Expression of glutamine synthetase genes, GS1, in the embryo. The increase in expression of the 6AL and 6BL homeologues in response to sulphur deficiency was significant (p = 0.0401 and 0.0164, respectively). (C) Expression of glutamate synthase (GOGAT) in the embryo. Expression of the D genome gene did increase in response to sulphur deficiency, and although the change was not statistically significant, it followed the trend for ASN2 and GS1. (D) Contrasting responses of asparagine synthetase gene ASN2 and the most highly expressed asparaginase gene in the endosperm. The increase in expression of all three asparaginase homeologues in response to sulphur deficiency was significant (p < 0.01), as was the decrease in expression of the ASN2 homeologues (p < 0.001 for both). (E) Expression of glutamate-cysteine ligase in the endosperm. The reduction in expression of the 1BS and 1DS genes in response to sulphur deficiency was significant (p < 0.001 and p = 0.028, respectively). Replotted from RNA-seq data provided in [52].