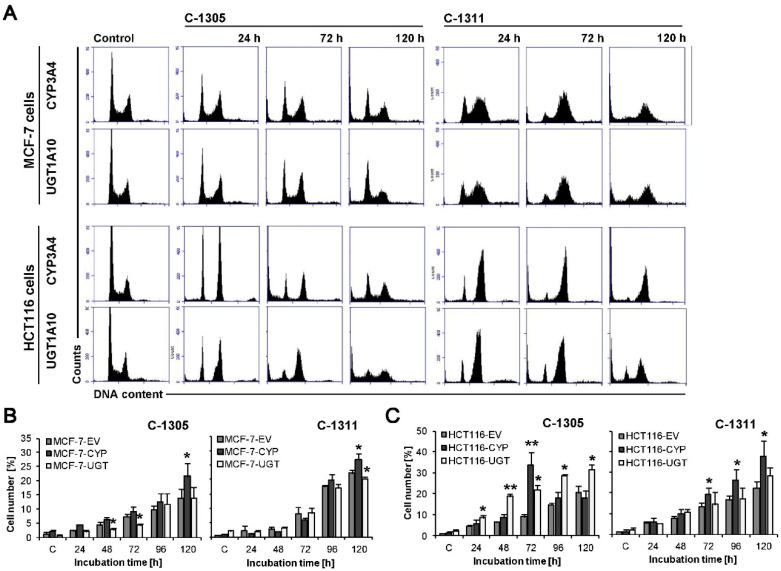
Figure 7.
Cell cycle distribution of MCF-7-CYP3A4, MCF-7-UGT1A10, HCT116-CYP3A4 and HCT116-UGT1A10 cells. Cells were untreated (control) or treated with C-1305 or C-1311 (IC80 for MCF-7 cells, IC90 for HCT116) for the times indicated and subjected to propidium iodide staining and flow cytometry as described in Materials and Methods. (A) Histograms show number of cells (y-axis) versus DNA content (x-axis) and are representative of at least three experiments for each condition. (B,C) Bar graphs show data quantitation with percentage of cells with less than 2N DNA (sub-G1 fraction): three types of MCF-7 (B) and HCT116 (C) cells treated with C-1305 or C-1311. Results are expressed as mean ± SEM (n ≥ 3). Significant differences in cell percentages between control, stably transfected with EV cells and UGT1A10- or CYP3A4-transfected are indicated as follows: * p ≤ 0.05; ** p ≤ 0.001.