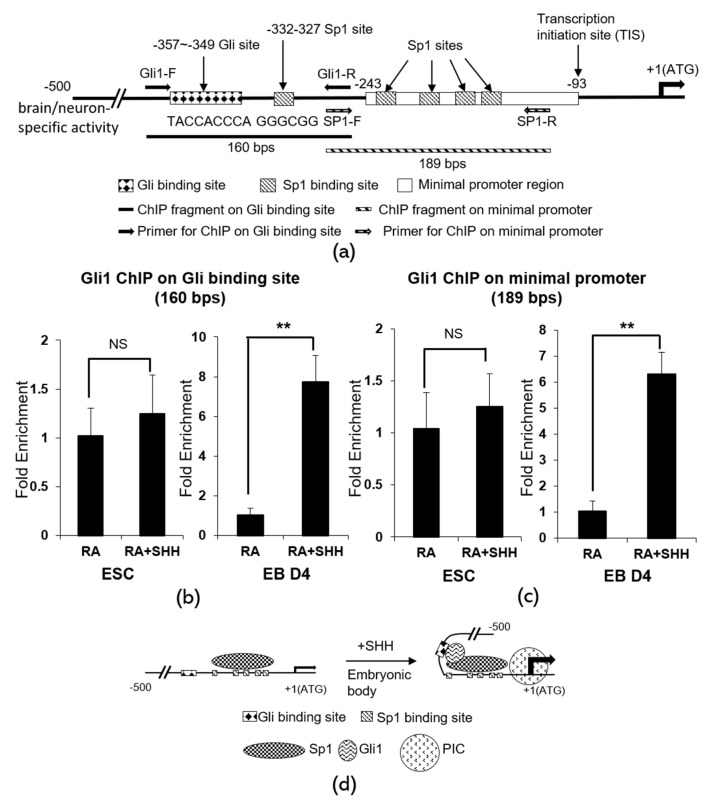
Figure 4.
ChIP assays for Shh activated Gli-induced chromatin conformational change on the Crabp1 promoter. (a) The map of neuron-specific promoter of Crabp1 gene and the design of ChIP assays. Gli1-F, forward primer for the Gli site-containing fragment; Gli1-R, reverse primer for the Gli site-containing fragment; SP1-F, forward primer for the minimal promoter fragment; SP1-R, reverse primer for minimal promoter fragment. The Gli-binding chromatin is indicated by 160 bps PCR fragment. The minimal promoter containing Sp1 sites is indicated by 189 bps PCR fragment. (b) ChIP results of Gli1 physical association with the Gli binding site (160 bps fragment) in embryo body (EB D4, right) vs. embryonic stem cells (ESC, left). (c) ChIP results of Gli physical association with the minimal promoter (189 bps fragment) in embryo body (EB D4, right) vs. embryonic stem cells (ESC, left). Results are presented as means ± SD; * p < 0.05, ** p < 0.01; compared RA+Shh to RA only group; NS, not significant. (d) A molecular model depicting Shh activation of Gli1-induced chromatin juxtaposition of the Gli-binding region with the Sp1-binding minimal promoter of Crabp1 gene. Change in this chromatin conformation brings the Gli1/Sp1 activator complex to the proximity of preinitiation complex (PIC), thereby facilitating transcription activation.