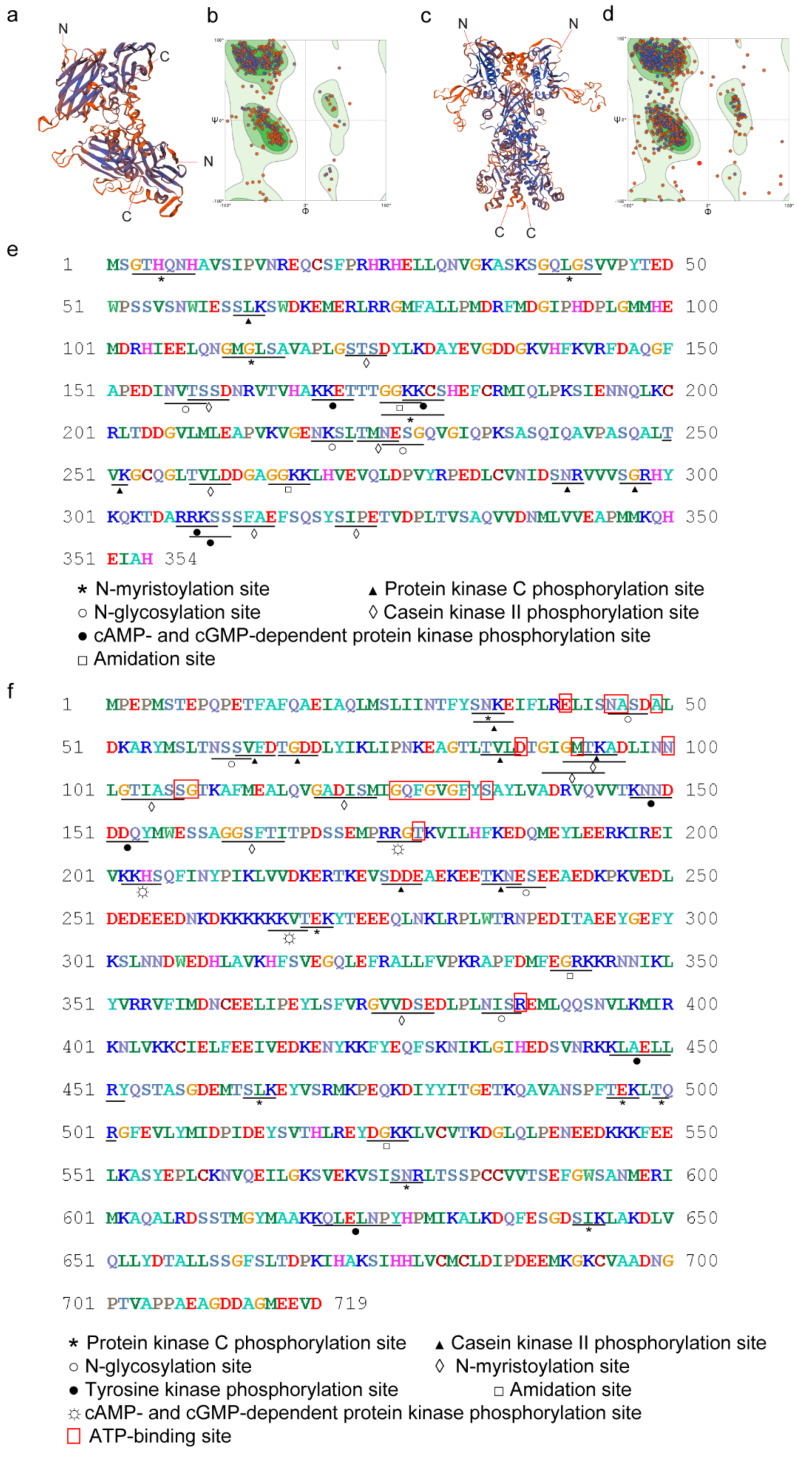
Figure 1.
Predicted 3D structure and motifs of Sjp40 and Sjp90α. Homology modeling was performed using the Swiss-Model Server and the predicted 3D structure of homo-dimer Sjp40 (a) and homo-dimer Sjp90 (c) are shown. Model quality was evaluated using the Ramachandran plot method and the results represent the acceptable stability of the 3D structure of Sjp40 (b) and Sjp90 (d). (e) Sjp40 motifs: *: N-myristoylation site; ▲: protein kinase C phosphorylation site; ◊: casein kinase II phosphorylation site; ○: N-glycosylation site; ●: cAMP- and cGMP-dependent protein kinase phosphorylation site; and □: amidation site. (f) Sjp90 motifs: *: protein kinase C phosphorylation site; ▲: casein kinase II phosphorylation site; ◊: N-myristoylation site; ●: tyrosine kinase phosphorylation site; □: amidation site; ☼: cAMP- and cGMP-dependent protein kinase phosphorylation site; and □: ATP-binding site.