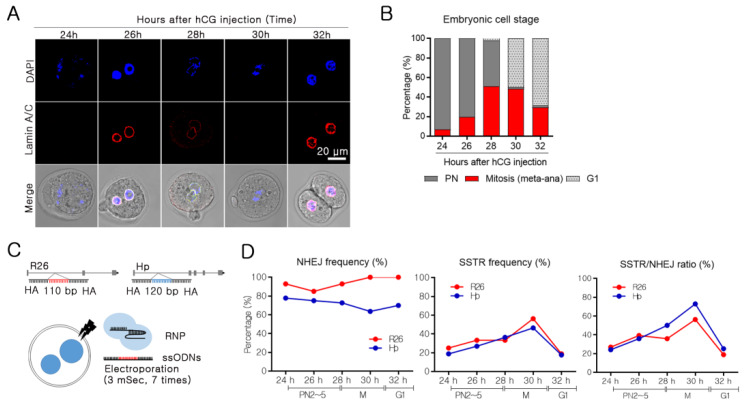
Figure 1.
Identification of embryonic cell cycle, and cell cycle correlation with non-homologous end joining (NHEJ) and single-strand template repair (SSTR) in mouse embryos. (A) For whole-mount staining of the nuclei (DAPI) (blue) and lamin A/C (red). Randomly divided embryos were used for immune staining at 24 h, 26 h, 28 h, 30 h, and 32 h after human chorionic gonadotropin (hCG) injection. Fluorescence was detected using a confocal microscope. The embryonic cell stage was defined based on the presence of a nuclear envelope, nuclear condensation, and embryonic division. A representative image is shown for each time point. (B) The embryonic cell stage was defined, and the frequency was calculated. Embryos with two pronuclei were designated to PN, embryos with condensing nuclei and without a nuclear membrane were considered as mitosis, and two-cell embryos were considered to be in G1 (Number of analyzed embryos after 24 h: 45, 26 h: 41, 28 h: 45, 30 h: 58, and 32 h: 44). (C) Schematic representation of small sequence insertion using single-stranded oligodeoxynucleotides (ssODNs) (target loci: Rosa26 and Haptoglobin (Hp)). For small sequence insertions of 110–120 bp into the target site, two binding site-overlapping sgRNAs and ssODNs with 40–45 nucleotides homology sequences were applied. (D) NHEJ frequency was calculated by PCR and T7E1 analysis after SpCas9 ribonucleoprotein (RNP) electroporation into embryos at each time point. The presence of SSTR was analyzed by single embryo PCR after SpCas9 RNP and ssODN electroporation, and the frequency was calculated as the % of KI embryos per total embryos.