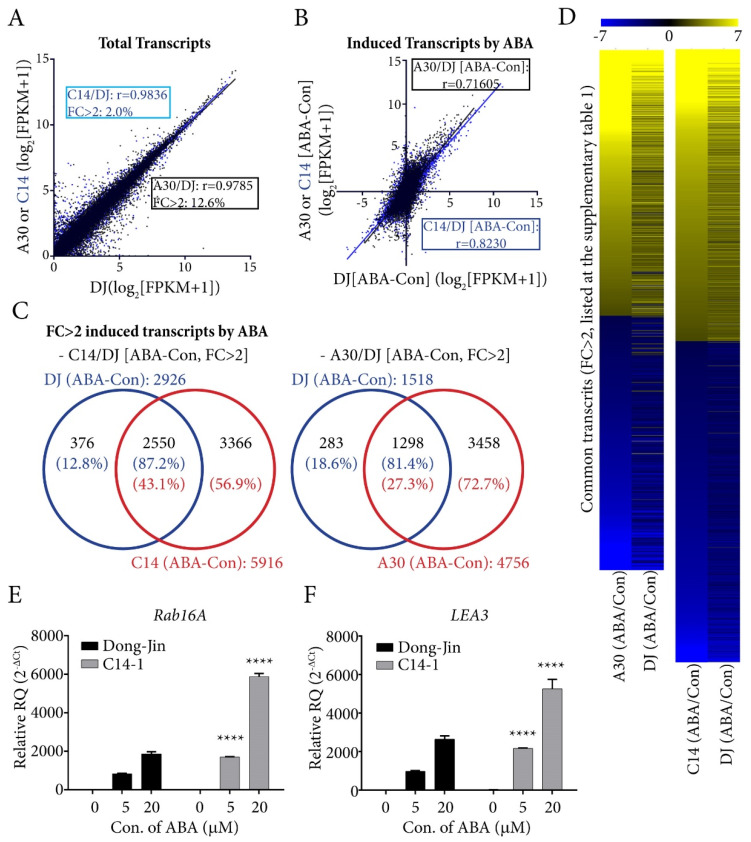
Figure 4.
Comparative transcriptome analysis of transgenic rice overexpresing OsPYL/RCAR7 and OsPYL/RCAR3. (A) Total transcripts of C14 (transgenic plants over-expressing OsPYL/RCAR7) (blue) and A30 (transgenic plants over-expressing OsPYL/RCAR3) (black) treated with no ABA were compared with DJ (wild type). ‘r’ is pearson correlation coefficient. FC is fold change. (B) Induced or reduced transcripts in C14 (blue) or A30 (black) treated with ABA were compared. (C) Venn diagram of transcripts with more than twofold changes under ABA treatment. (D) Heat map of common trascripts (fc > 2). (E,F) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis with ABA dependently induced gene. Each transcript was detected in treated samples with indicated ABA treatment for 12 h. Similar expression levels were observed in triplicates. Error bars are SD. **** means that p-value is less than 0.0001. FPKM, fragment per kilo base of transcript per million mapped reads.