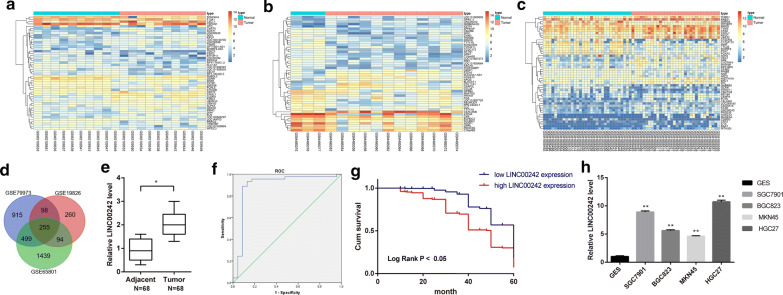
Fig. 1.
Expression pattern of LINC00242 in GC tissues and cells. a–c, Heat maps depicting the DEGs obtained from the GC-related datasets GSE79973, GSE19826 and GSE65801. The abscissa represents the sample number, and the ordinate represents the differentially expressed gene; the left dendrogram represents gene expression cluster; each rectangle corresponds to a sample expression value; the histogram at the upper right refers to color gradation. d Venn analysis of DEGs in GC microarray, three circles in the figure respectively represent the DEGs from the three datasets, and the middle part represents the overlaps. e The expression of LINC00242 in 68 pairs of GC and adjacent normal gastric tissues was determined by RT-qPCR. f The area under the ROC curve according to LINC00242 expression. The threshold of LINC00242 expression was 0.803. g The Kaplan–Meier method was used to plot GC patient survival according to LINC00242 expression. f The expression of LINC00242 in GES, SGC7901, BGC823, MKN45 and HGC27 cell lines was determined by RT-qPCR. Results are presented as mean ± S.D. of three technical replicates. *(compared with adjacent normal gastric tissues) indicates p < 0.05 by paired t-test. ** (compared with GES cells) indicates p < 0.01 by one-way ANOVA followed by Tukey’s test