Figure 7.
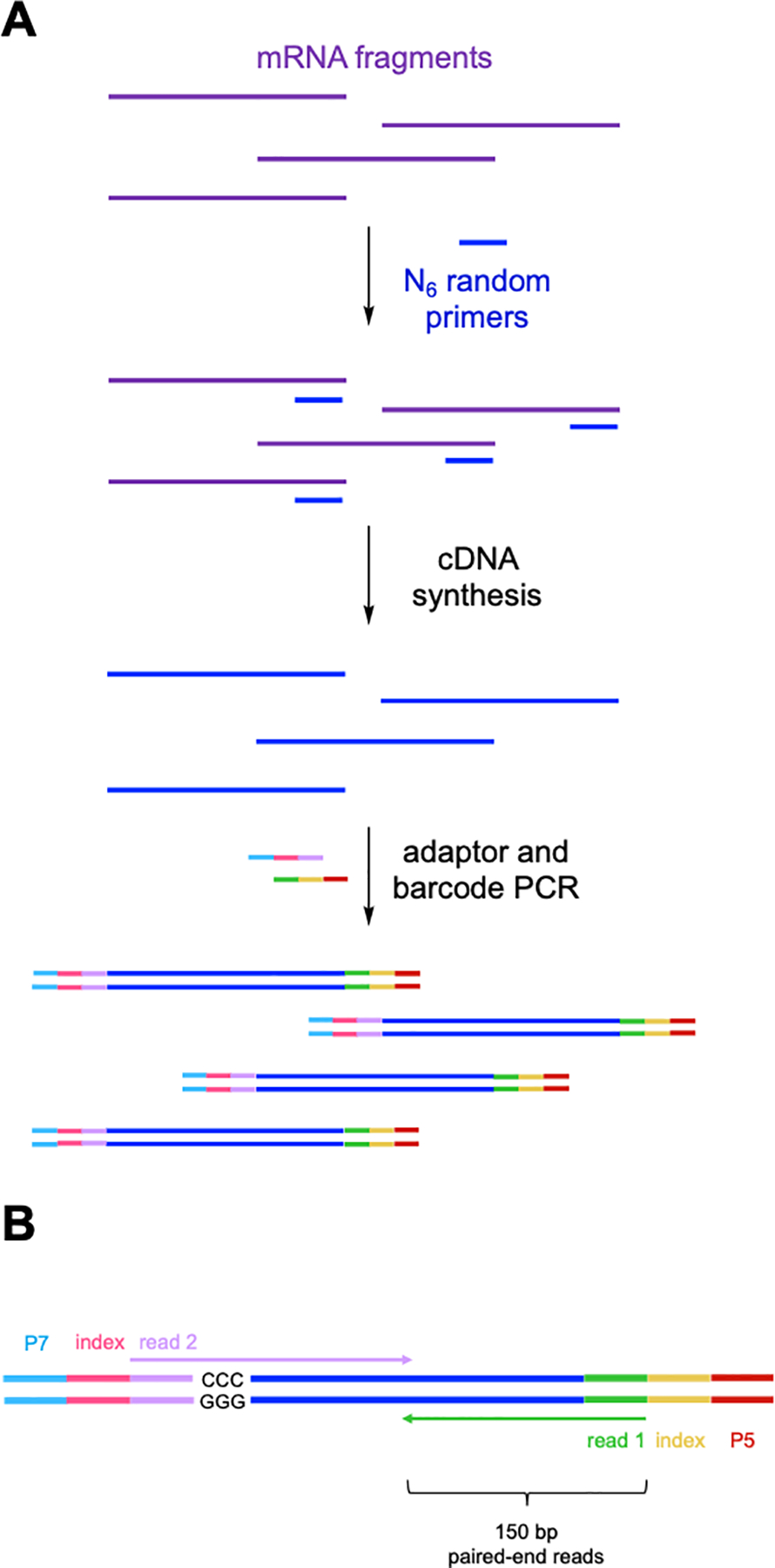
(A) Key steps in SMARTer® RNA-seq library preparation following EndoVIPER pulldown. Enriched mRNA fragments are first reverse-transcribed using random hexamer (N6) DNA primers. Resulting cDNA is then PCR amplified to attach (B) Illumina flow cell adaptors (P7 and P5), Illumina TruSeq® index barcodes, and read primer binding sites (reads 1 and 2). 150 bp paired-end sequencing is then used to ensure coverage of the insert space from both reads. The template switching oligo step produces “GGG/CCC” during cDNA synthesis. These must be removed from the beginning of read 2 during data analysis.
