Figure 3.
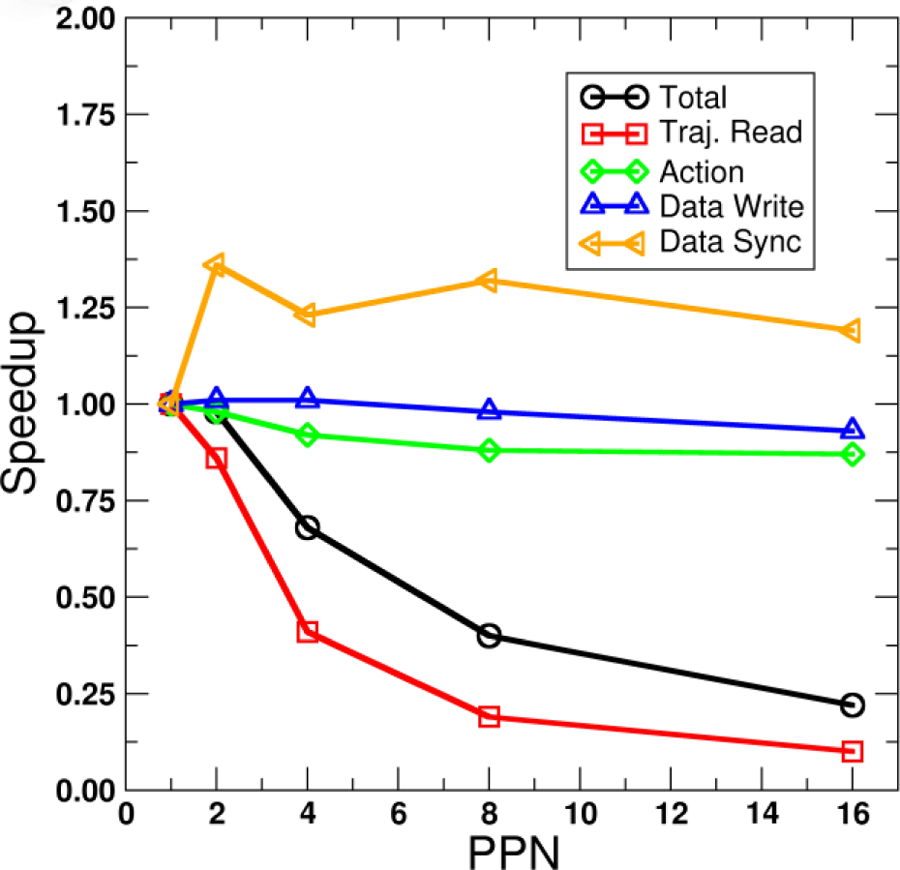
Speed up versus number of MPI processes per node (PPN) for across-trajectory parallelization of a hydrogen bond calculation (375 acceptor atoms, 371 acceptor/donor sites, 440 solute hydrogens, 18000 frames) using 16 MPI processes. Each line represents timings for different parts of the trajectory processing Run (total time, trajectory read, the hydrogen bond action, data file write, and data set sync). Raw timings for 1 PPN (16 nodes): Total=22.62±0.86 s, Traj. Read=8.27±1.12 s, Action=2.35±0.06 s, Data Write=9.14±0.06 s, Data Sync=0.08±0.01 s. Calculations run on the LoBoS cluster, NIH (Haswell 2.4 GHz dual socket eight-core nodes, NFS-mount file storage).
