Graphical abstract
Keywords: Graphitic carbon nitride QDs, Magnetoplasmonic molybdenum trioxide QDs, Influenza virus, Fluoroimmunosensing, Localized surface plasmonic resonance
Highlights
-
•
Magnetic/plasmonic-assisted fluoro-immunoassay system is developed for virus detection.
-
•
Magnetic-derivatized plasmonic molybdenum trioxide quantum dots (MP-MoO3 QDs) is the plasmonic/magnetic agent.
-
•
Fluorescent graphitic carbon nitride quantum dots (gCNQDs) Is the monitoring probe.
-
•
Vrus was formed a nano-sandwich between the antibody-conjugated nanomaterials (Ab-MP-MoO3 QDs and Ab-gCNQDs.
-
•
This interaction resulted in the modulation and gradual enhancement of the photoluminescence intensity.
Abstract
A novel magnetic/plasmonic-assisted fluoro-immunoassay system is developed for the detection of influenza virus using magnetic-derivatized plasmonic molybdenum trioxide quantum dots (MP-MoO3 QDs) as the plasmonic/magnetic agent and fluorescent graphitic carbon nitride quantum dots (gCNQDs) as the monitoring probe. Specific antibody against influenza A virus was conjugated onto the surface of MP-MoO3 QDs and gCNQDs, respectively. In the presence of influenza A virus (as the test virus), a core-satellite immunocomplex is formed between the antibody-conjugated nanomaterials (Ab-MP-MoO3 QDs and Ab-gCNQDs) and their interaction resulted in the modulation and gradual enhancement of the fluorescence intensity of the detection probe with the influenza virus concentration-dependent increase. In addition, PL change without influenza A virus was not observed. Limits of detection of 0.25 and 0.9 pg/mL were achieved for Influenza virus A/New Caledonia (20/99/IVR/116) (H1N1) detection in deionized water and human serum, respectively. Clinically isolated influenza virus A/Yokohama (110/2009) (H3N2) was detected in the range of 45 – 25,000 PFU/mL, with a limit of detection ca 45 PFU/mL (as opposed to a minimum of 5000 PFU/mL for a commercial test kit). This developed biosensor provides a robust, sensitive as well as a selective platform for influenza virus detection.
1. Introduction
The recent outbreak of a novel coronavirus disease (COVID-19) and its potential for adverse effect on the global economy is an example of how quickly new infectious diseases can arise and spread [1]. Outbreaks such as this can unexpectedly cause demands for clinical knowledge, rapid diagnostic strategies, and epidemiological studies before a pandemic occurs. The rapid and ultrasensitive detection of infectious diseases is critical for the prevention and/or control of outbreaks. Therefore, it is crucial to continually deploy innovative materials to further develop the practical applications of biosensing systems capable of detecting biomolecules (DNAs, RNAs, proteins and virus particles) and of diagnozing other potentially harmful infectious diseases.
The deployment of carbon-based quantum dots as optical probes has received a tremendous boost due to their excellent optical properties, biocompatibility and low cost of preparation [[2], [3], [4], [5], [6], [7]]. As carbon-based nanomaterials, graphitic carbon nitrides QDs (gCNQDs) possess excellent optical properties comparable to traditional heavy metal-based QDs, and are not as toxic. So, they are becoming competitive materials in nanosensors development [[8], [9], [10], [11]]. The presence of graphitic nitride-N-atoms introduces a different kind of “surface state” which impacts some semiconductor-like properties [[12], [13], [14]]. This feature has attractively bestowed an edge on gCNQDs and their derivatives for utilization in optical and electrochemical-based sensing [15,16]. For instance, a robust photo-electrochemical immunosensor was designed by Sun et al. for the sensitive detection of avian viruses using hybrid of gold nanoparticles (AuNPs) and gCNQDs coupled to CdTe QDs [10]. Wang et al. reported a photo-electrochemical platform for methylated RNA detection using gCNQDs/CdS hybrid [17]. Pang et al. reported the fabrication of pcDNA3-HBV nanobiosensor using gCNQDs-sensitized TiO2 nanopillars [9]. Interestingly, the tailored surface modification of gCNQDs has been achieved with moieties that acted as receptors to capture and detect target analytes. In our previous work, the surface of gCNQDs was functionalized with 2, 2, 6, 6-tetramethyl (piperidin-1-yl) oxyl (TEMPO) to detect ascorbic acid in the presence of zinc phthalocyanine [2]. Furthermore, biomolecules including thymine and tannic acid have been grafted onto the surface gCNQDs for the purpose of deriving Hg2+, Cu2+ and ascorbic acid responsive nanosensors, respectively [18,19].
On the other hand, novel materials which can serve as viable alternatives to Au and Ag are desired and are a hotspot for research [20,21]. This is due in large part to the high cost of preparation and/or procurement of noble metals nanoparticles (NPs). As a result, a new low-cost, easy-to-prepare and non-toxic molybdenum trioxide QDs (MoO3 QDs) with excellent plasmonic properties are gaining traction [[21], [22], [23], [24], [25], [26], [27], [28]]. MoO3 QDs are few-crystalline-structured nanoparticles with single-layered morphology and oxygen vacancies. Hence, they exhibit a semiconductor-based tunable localized surface plasmonic resonance (LSPR), comparable to noble metals NPs, both in the visible and near-infrared (NIR) regions [27,28]. To demonstrate that the inherent tunable LSPR of MoO3 QDs can be harnessed for opto-electrical sensing, MoO3 QDs was adopted as a substrate for the surface enhanced Raman spectroscopy (SERS) detection of bovine serum albumin (BSA) and methylene blue (MB), respectively. The Raman signals of the probe molecule (Rhodamine 6 G) and MB were strongly amplified due to the interfacial charge transfer effect between the substrates and probe molecule. The intense LSPR absorption of MoO3 QDs in the near infrared (NIR) region was deployed in the photothermal ablation of cancer, glucose detection in the fluorescence (FL)-based detection of 2, 4, 6-trinitrotoluene (TNT) [23,25].
Plasmonic nanostructures are known to influence the fluorescence (FL) properties of QDs via plasmon-induced energy transfer [29,30]. Optical biosensing platforms have been designed based on this kind of interaction [31,32,33,34,35]. For the first time, we have synthesized novel, water-soluble magnetoplasmonic MoO3 QDs and examined their potential as a plasmonic material in the presence of gCNQDs for ultrasensitive FL signal enhancement-based assay to detect extremely low concentrations of influenza virus A (H1N1) and (H3N2) RNAs. The use of gCNQDs/MP-MoO3 QDs as a novel and interesting combination of functional materials for optical biosensing was inspired by the high FL of gCNQDs, the strong plasmonic effect of MP-MoO3 QDs, their intrinsic non-toxicity and the desire to fabricate low-cost biosensing systems. As a result, a simple and sensitive fluoroimmunoassay of influenza virus was developed using hybrid combination of gCNQDs and MP-MoO3 QDs. The detection principle was based on the antigen-antibody interaction achieved between the immunocomplex of antibody-conjugated gCNQDs and MP-MoO3 QDs, respectively. The detection protocol maintained a magnetic separation and purification of the target influenza virus from complex matrices and impurities owing to the magnetic functionality of the MoO3 QDs which was another extremely useful feature apart from plasmonic effects. This was intended to optimize the FL signal enhancement of the probe QDs. This step, as expected, resulted in a signal enhancement and sensitivity of the established immunoassay. The target influenza virus was separated easily by an external magnet field allowing an ultrasensitive detection of influenza virus A (H1N1) and clinically isolated influenza virus A (H3N2) RNA, respectively. The biosensing system has been developed to provide a robust performance, as well as high selectivity and ultrasensitivity for influenza virus detection when compared to a commercially available rapid influenza diagnostic test (RIDT) kits.
2. Materials and methods
2.1. Materials
Melamine, glutaraldehyde, citric acid (CA), dimethyl formamide (DMF), iron (II) chloride, iron (III) chloride, bovine serum albumin (BSA), N-hydroxysuccinimide (NHS), and N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich (St Louis, USA). Polyvinylpyrrolidone (PVP), ammonium molybdate tetra-hydrate were supplied by Wako Pure Chemicals Ind. (Osaka, Japan).
Anti-human influenza A (H1N1) monoclonal antibody (clone C179), anti-human influenza A (H3N2) monoclonal antibody (Clone F49) and anti-human influenza A (H1, H2, H3) monoclonal antibody (Clone C111) which is positive for both influenza viruses H1N1 and H3N2 was purchased from Takara Bio. Inc, (Kusatsu, Shiga, Japan). Influenza virus A/New Caledonia (20/99/IVR/116) (H1N1) was purchased from ProSpec-Tany TechnoGene Ltd. (Rehovot, Israel). Clinically isolated influenza virus A/Yokohama/110/2009 (H3N2) was kindly provided by Dr. C. Kawakami of Yokohama City Institute of Health, Japan. Goat anti-rabbit IgG-HRP were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). Commercial RIDT kit - QuikNavi Flu 2 was purchased from Denka -Seiken Co. Ltd. (Tokyo, Japan). Zika virus was kindly provided by Prof. K. Morita of Institute of Tropical Medicine, Nagasaki University. Noro virus-like particles (NoV-LPs) were prepared in our lab according to previously reported protocol [36]. HEV-LPs were prepared according to Li et al. [37]. All experiments were carried out using high purity deionized (DI) water (>18 MΩ cm).
2.2. Equipment
Ground state electronic absorption (UV/Vis), fluorescence excitation and emission spectra were recorded on a filter-based multimode microplate reader (Infinite F200 M; TECAN, Ltd, Männedorf, Switzerland). Images of transmission electron microscope (TEM) and scanning electron micrograph (SEM) were obtained using JEM-2100 F (JEOL, Ltd., Tokyo, Japan) operating at 100 kV and 200 kV, respectively. The morphological and structural characteristics of gCNQDs were also obtained using a high resolution transmission electron microscope (HRTEM) coupled with a JEOL 2010 TEM operating at an accelerating electrical potential of 200 kV.
Atomic force microscopy (AFM) measurement in tapping mode was carried out with MFP-3D Origin supplied by Asylum research (Oxford instruments company, USA). AFM analysis was done by drop casting and room temperature drying of the samples solutions on a freshly cleaved mica surface. Dynamic light scattering (DLS) and zeta potential experiments were done on a Malvern Zetasizer nanoseries, Nano-ZS90 (Malvern Inst. Ltd., Malvern, UK). Powder X-ray diffraction (PXRD) analysis was carried out using a RINT ULTIMA XRD (Rigaku Co., Tokyo, Japan) with αNi filter and a Cu-Kα source. Data were collected over 2θ = 30 – 90° at a scan rate of 0.01°/step and 10 s/point. Fourier transform infrared spectroscopy was performed using FT/IR-6300 with ATR PRO610P-S (JASCO, Tokyo, Japan). Raman spectroscopic measurements were carried out using NRS-7100 Raman Spectrometer with f500 spectrograph (JASCO, Tokyo, Japan). Conjugation of the antibody to the respective QDs and nanoparticles was confirmed by enzyme-linked immunosorbent assay (ELISA) using a microplate (Model 680; Bio-Rad, Hercules, USA).
2.3. Synthesis of graphitic carbon nitride QDs (gCNQDs)
Melamine was used to synthesize graphitic carbon nitride nanosheets (gCNNs) according to reported procedures [38]. Graphitic carbon nitride QDs (gCNQDs) were prepared by the solvothermal treatment of the graphitic nanosheets (gCNNs) according to previously reported method with modifications [18]. Briefly, gCNNs (0.1 g) and citric acid (0.2 g) were dissolved in 10 mL of DMF, stirred for 5 min and sonicated for 20 min to obtain homogenous suspension. The suspension was transferred and sealed in a 50 mL Teflon-lined stainless steel autoclave and heated at 160 °C for 12 h. The autoclave was allowed to cool to room temperature naturally. The obtained product was filtered through a 0.22 μm microporous filter membrane and then dialyzed (using a membrane of MWCO 1.5 kDA) for 48 h to obtain pure gCNQDs solution. The solution was further freeze dried to obtained solid product.
2.4. Synthesis of magnetoplasmonic molybdenum trioxide QDs (MP-MoO3 QDs)
Pristine MoO3 QDs was prepared by a room temperature ultraviolent (UV) irradiation method according to reported procedure [28]. The novel magnetic-derivatized MP-MoO3 QDs were synthesized under hydrothermal conditions. In a typical experiment, 0.15 g (0.12 mmol) of ammonium molybdate tetrahydrate was dissolved in 20 mL mixture of deionized water and HCl (9:1). Then 0.1 g of PVP was added to the mixture and stirred vigorously. After 5 min, 1 mmol of FeCl2 was added and the mixture was subsequently irradiated under a 365 nm UV light with constant stirring. A dark blue product of MoO3 QDs was formed after 30 min and was left for a total of 1 h. The product was centrifuged to remove unreacted/excess PVP and dialyzed using a 2.0 kDA dialysis tubing membrane. Next, 5 mmol of FeCl3 was added to 10 mL of the obtained product and transferred to a Teflon-line stainless steel autoclave and heated at 160 °C for 4 h. Subsequently, the product was washed with ethanol by centrifugation and further purified by magnetic separation. To surface functionalize the MP-MoO3 QDs with carboxyl groups, 2 mL of glutaraldehyde was incubated with 2 mg of MP-MoO3 QDs in ethanol stirred for 12 h. The product was purified using magnetic separation. All experiments were carried out using ultrapure DI water.
2.5. Conjugation of antibodies to gCNQDs and MP-MoO3 QDs
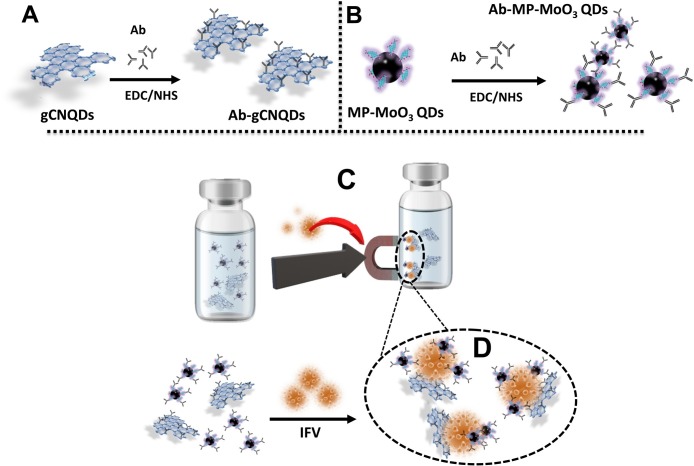
Anti-human influenza virus A (H1N1) (Clone C179) or (H1, H2, H3) (Clone C111) monoclonal antibody was conjugated to gCNQDs via the well-known EDC/NHS covalent chemistry (Scheme 1 A). Briefly, 100 μL of 0.1 M EDC was added to 2 mL of gCNQDs solution to activate the carboxylic groups on their surfaces. The solution was stirred at ambient temperature for 30 min. Next, 100 μL of 0.1 M NHS was added to the mixture and further stirred for ∼15 min followed by the addition of 5.1 μg/mL of the antibodies in PBS 7.6. The resulting mixtures were then stirred for 8 h at 7 °C. The Ab-conjugated gCNQDs were purified by centrifugation (3000 × g, 5 min) and subsequently dissolved in 2 mL of ultrapure water and stored in the refrigerator for further use. Anti-human influenza virus A (H1N1) monoclonal antibody (Clone C179) was conjugated to MP-MoO3 QDs following the above described procedures for the detection of influenza virus A/New Caledonia/20/99/IVR/116 (H1N1) as shown in Scheme 1B. Anti-human influenza virus A (H3N2) monoclonal antibody (Clone F49) was conjugated to MP-MoO3 QDs for the detection of the clinically isolated influenza virus A/Yokohama/110/2009 (H3N2). The conjugation of antibodies (Ab) to gCNQDs and MP-MoO3 QDs, respectively, was confirmed using ELISA. Details have been provided in our previous work [33].
Scheme 1.
(A) Antibody conjugation to gCNQDs via EDC/NHS chemistry. (B) Antibody conjugation to MP-MoO3 QDs via EDC/NHS chemistry. (C) Magnetic separation and purification step upon target virus addition (D) Core-satellite immunocomplex of gCNQDs and MP-MoO3 QDs in the presence of influenza virus.
2.6. Immunofluorescence detection of influenza virus (H1N1 or H3N2)
The respective antibodyconjugated gCNQDs and MP-MoO3 QDs were used as the detection probe (5% BSA was used for blocking to avoid non-specific binding of the antibody-conjugated probes). Next, 50 μL of the probe solution was placed in a 96-well plate, followed by the incubation of the respective target influenza virus (20 μL) for 5 min before fluorescence measurements were acquired. After the incubation duration, the antibody-antigen complex solutions were excited at 500 nm, and the fluorescence spectra within the range of 530–800 nm were recorded. The detection of influenza virus (H1N1) was carried out in ultrapure water and human serum within the concentration range of 1 pg/mL – 100 ng/mL. Clinically isolated H3N2 RNA was detected within the range of 45 – 25,000 PFU/mL. All detection experiments were done in triplicate. The limits of detection (LODs) were evaluated by using 3δ/K0, where δ is the standard deviation of blank measurement (n = 12) and K0 is the slope of the generated calibration curves following replicated measurements [39].
3. Results and discussion
3.1. The design of the biosensing platform
The oxygen-deficient MoO3 nanostructures, like Au NPs, display intensive LSPR absorption in the NIR-I region [27,28]. The absorption spectra of MoO3 QDs strongly overlap with the FL spectra of gCNQDs (Fig. S1 in Supplementary data). The beneficial NIR plasmonic absorption of MoO3 QDs was therefore used in enhancing the FL signal of gCNQDs via a plasmonic-induced energy transfer process resulting from their close proximity in a core-satellite network triggered by the “virus-antibody” affinity (Scheme 1C, D and Fig. S2 in Supplementary data). In addition, MoO3 QD's magnetic derivatization was extremely useful in the purification the antibody-QDs conjugates produced and in the isolation of the influenza antigens specifically captured, thus increasing the sensitivity of the developed immunoassay. The immobilization of anti-influenza virus antibodies on the surfaces of gCNQDs and MP-MoO3 QDs ensured the successful and precise binding of the antibody-conjugated materials to influenza virus. The extent to which gCNQDs interact with MP-MoO3 QDs to enable the plasmonic-induced effect was dependent on influenza virus concentration. When the concentration of influenza virus is increased, more plasmonic MP-MoO3 QDs are propelled closer to the fluorescent gCNQDs which triggered the spontaneous enhancement in the FL of the gCNQDs.
3.2. Structural/optical characterization of graphitic carbon nitride QDs
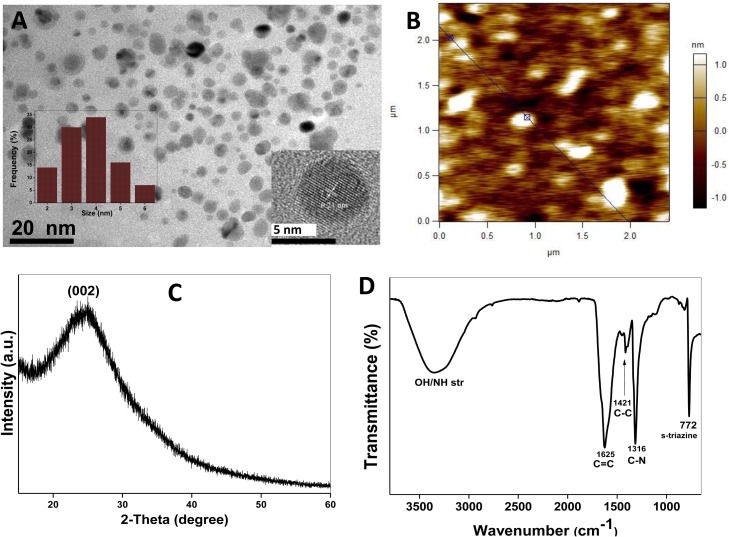
In this work, bulk graphitic carbon nitride nanosheets (gCNNs) were treated under solvothermal conditions to prepare highly fluorescent gCNQDs. In order to characterize the prepared carbon-based QDs, various techniques ranging from materials science to spectroscopy were employed. The transmission electron microscopy (TEM) image obtained showed quasi-spherically shaped particles that are monodispersed with size distribution between 5–9 nm (7.25 ± 0.7 average diameter) (Fig. 1 A). The HRTEM image shows that the graphitic lattice spacing is 0.23 nm (Fig. 1A inset), which is a feature of the (002) hexagonal plane of graphitic carbon nitrides [4,8,11]. The AFM results showed a narrow distribution of the graphitic QDs with few layers of planar graphitic sheets having nanoscopic size of ∼5 nm with a height profile of 1.5 nm (Fig. 1B). The powder X-ray diffraction (XRD) pattern (Fig. 1C) obtained for the prepared gCNQDs exhibit a broad peak at 27.9° which corresponds to the graphitic interplanar 002 d-spacing which is known to be exhibited by graphitic QDs [8]. Chemical functional groups were also elucidated by carrying out FTIR spectroscopy. In Fig. 1D, the spectra of gCNQDs displayed typical vibrations inherently possessed by the precursors (gCNNs/CA). An intensely broad absorption was observed at ∼3688 – 2792 cm−1 which is due to the combined stretching vibrations of N—H and O—H (from carboxyl group). The intense bands appearing at 1431, 1316 and 772 cm−1 are reminiscent of the stretching modes of C—C, C—N and breathing mode of the s-triazine heterocycles characteristic of carbon nitrides [4,8,11]. The C O observed at 1625 cm−1 confirms the presence of carboxylic groups. To complement the surface functional groups elucidation results obtained from FTIR analysis, zeta potential analysis was performed in order to understand the stability of the gCNQDs in solution and their surface chemistry. The results obtained show that the zeta potential of the gCNQDs was −18.1 mV (Fig. S3A in Supplementary data), which suggests that some carboxylic (COOH) functionality are present on the gCNQDs surface. In comparison to some reported carbon dots (CDs) and graphene QDs [40], the lower negative zeta potential value may indicate the existence of amino (NH2) and/or nitride functionality of the s-triazine heterocycles expected to be associated with graphitic carbon nitride QDs [41]. The result also shows that the gCNQDs may be dispersed in solution to a moderate degree, as zeta potential values >20 mV are known to result in well-dispersed colloidal solutions due to increased interparticle repulsion [40,41].
Fig. 1.
Characterization results of gCNQDs showing (A) TEM image (inset HR-TEM). (B) AFM (C) XRD pattern and (D) FTIR spectra.
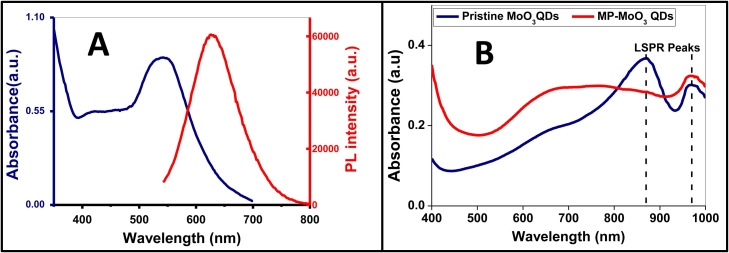
The optical spectroscopy characterization of gCNQDs displayed a typical strong ground-level absorption peak at ∼542 nm and a broad absorption at < 500 nm (Fig. 2 A). These absorptions are ascribed to the n-π* and π-π* electronic transitions of the electron lone pairs of N atoms of s-triazine units [4,8,11,18]. Further, the fluorescence properties of the as-synthesized gCNQDs were probed. The FL emission of gCNQDs prepared under solvothermal conditions exhibit emission extending into the yellow region with maximum fluorescence intensity at 652 nm when excited at 500 nm wavelength (Fig. 2A). Carbon-based QDs with similar fluorescence emission extending well into the red region have been prepared using solvothermal processes [42,43]. Another important parameter evaluated for the prepared gCNQDs in terms of their suitability to function as an optical probe is their fluorescence quantum yield (ΦF). The robustness and sensitivity of a given fluorescence-based material as a probe is dictated by efficiently high ΦF values. Interestingly, gCNQDs possess ΦF of ∼44 %; using Rhodamine 6 G as the reference standard (see Supplementary Information for details). It is plausible to infer that the presence of citric acid made the gCNQDs water soluble (due to the incorporation of carboxylic group), and also created additional defects/trap sites on the QDs surface (new surface states) [44,45]. This probably may have resulted in an easy exciton mobility translating into such a high ΦF and red-shifted emission.
Fig. 2.
(A) UV–vis and FL emission of gCNQDs. (B) The UV–vis absorption spectra of MoO3 QDs and their magnetic derivative. Solvent: ultrapure water. λex(gCNQDs) = 500 nm.
3.3. Characterization of magnetoplasmonic MoO3 QDs (MP-MoO3 QDs)
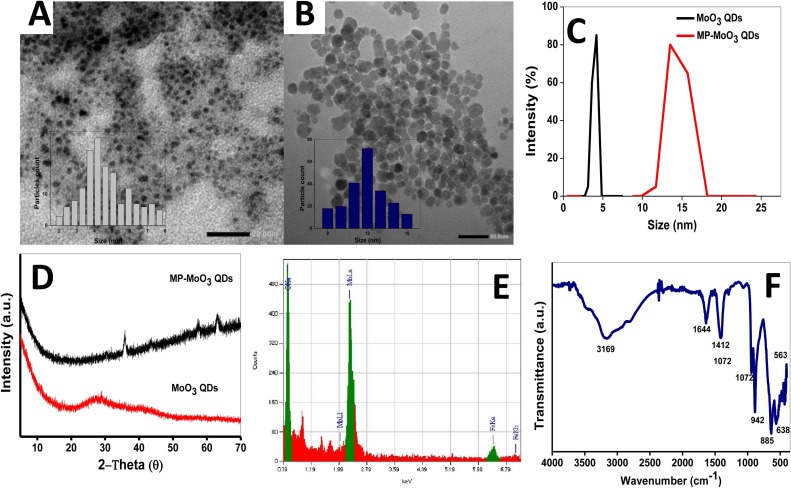
Novel magnetoplasmonic derivative of molybdenum-based QDs was prepared in the presence of FeCl2 and FeCl3 as the magnetic component precursors. FeCl2 was firstly reduced in situ by PVP and the subsequent hydrothermal treatment in the presence of FeCl3 ensured the surface grafting with magnetic NPs. Thanks to the strong reducing and stabilizing properties of PVP, a dual role was played to achieve the facile synthesis of MP-MoO3 QDs hybrid. PVP acted as a surface stabilizer/reducing agent for the magnetic NPs formation and initiated a strong plasmonic resonance [28,46]. Hence, the prepared MP-MoO3 QDs manifested an excellent aqueous solubility, stability and monodispersity. Optical characterization revealed that the UV–vis absorption of the pristine MoO3 QDs and their magnetic derivative (MP-MoO3 QDs) displayed LSPR peaks in the NIR with a slight blue shift and peak broadening observed for the MP-MoO3 QDs (Fig. 2B). To probe the morphology of the MP-MoO3 QDs, transmission electron micrograph (TEM) images were acquired for the pristine MoO3 QDs (without magnetic NPs) and MP-MoO3 QDs, respectively, (Fig. 3 A and B). A relatively narrow size distribution with near uniform shape was observed for MP-MoO3 QDs with an average size of 13.2 ± 0.5 nm. The pristine MoO3 QDs possess average size of ∼5 nm with a quasi-spherical morphology. Similar to TEM, a size increase was observed in DLS analysis for the prepared hybrid. The mean hydrodynamic diameter of the pristine MoO3 QDs at ∼3.2 nm was increased to ∼18 nm upon magnetic NPs functionalization (Fig. 3C). X-ray diffractogram (XRD) patterns provided an insight into MP-MoO3 QDs formation when compared with the pattern of the pristine MoO3 QDs. As shown in Fig. 3D, pristine MoO3 QDs are endowed with a broad peak within the range of 22 – 33°, which shows that they have poor crystallinity [27,28]. This peak has been attributed to the (040) positions of a-MoO3 (JCPDs no. 05-0508) [27]. Conversely, the MP-MoO3 QDs hybrid displays the characteristic diffraction pattern of MoO3 QDs in 2θ range from 5 – 30° and that of magnetic constituent at 2θ = 35°, 57° and 63° which corresponds to magnetic NPs marked indices of (311), (511) and (440), respectively [47]. Energy dispersive X-ray spectroscopy (EDX) was employed to elucidate the elemental compositions of MP-MoO3 QDs. The main compositional elements which are Mo and Fe are found as displayed in the obtained spectra (Fig. 3E). The characteristic infrared absorptions which have been assigned to Mo-O-Mo stretching vibrations in MoO3 QDs were clearly observed in the as-synthesized MP-MoO3 QDs in the region within 500 – 885 cm−1. Mo = O stretching vibration was observed at 942 cm−1 (Fig. 3F) [27]. Vibrations corresponding to the moieties of PVP as the surface stabilizing agent are further observed at 3169, 1644 and 1412 cm−1 (Fig. 3F). The acquired Raman spectra for pristine MoO3 QDs show the absorption typical of the Mo3-O and Mo2-O vibrations at 776 and 820 cm−1, respectively [27]. Compared to MP-MoO3 QDs, similar absorption occurred with the emergence of new peaks (Fig. S4 in Supplementary data), possibly due to the surface functionalization with magnetic NPs. These results indicate the successful preparation of the novel MP-MoO3 QDs nanocomposite. Further, the antibody conjugated MP-MoO3 QDs (Ab- MP-MoO3 QDs) or Ab-gCNQDs were subjected to DLS analysis in order to determine their hydrodynamic sizes. As shown in Fig. S5 in Supplementary data, Ab-gCNQDs show an average hydrodynamic size increase from an average size of ∼7.5 nm to ∼20 nm (Fig. S5A in Supplementary data). In a similar manner, the size recorded for Ab-MP-MoO3 QDs was ∼24 nm showing some increase from ∼ 13 nm due to the antibody conjugation, Fig. S5B in Supplementary data, thus confirming their functionalization, respectively.
Fig. 3.
Characterization of MP-MoO3 QDs showing (A) TEM image of pristine MoO3 QDs (B) TEM of magnetoplasmonic MoO3 QDs. (C) DLS, (D) XRD patterns, (E) EDX and (F) FTIR spectra.
3.4. Magnetoplasmonic-amplified detection of influenza virus (H1N1)
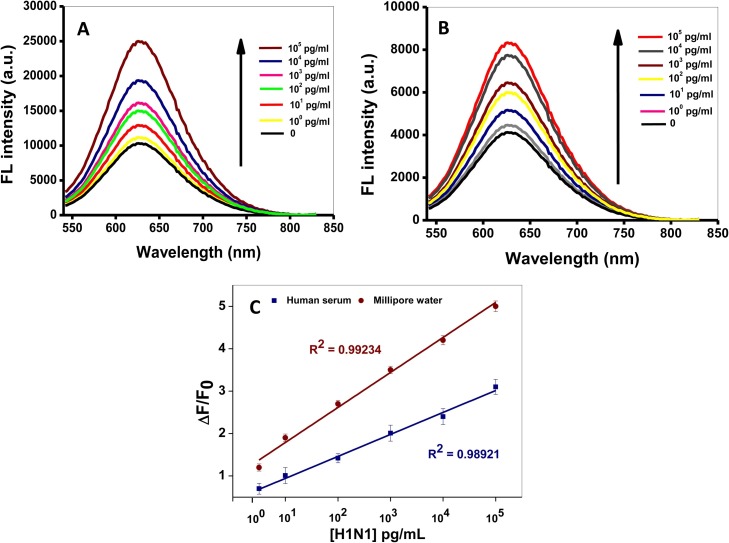
The virus detection protocol was established by using commercially obtained pure influenza virus (H1N1). Human serum medium was used to mimic complex biological media so as to demonstrate the applicability of the fabricated probe for influenza virus detection in clinical samples. Thus, gCNQDs was deployed as the fluorescent signal reporter and MP-MoO3 QDs as the plasmonic and magnetic material. MP-MoO3 QDs induced a steady and gradual plasmonic-induced enhancement of the FL of gCNQDs in the presence of increasing concentrations of influenza virus (H1N1) in water and human serum, respectively (Fig. 4 A and B). The core-satellite immunocomplex formed between the Ab-gCNQDs and Ab-MP-MoO3 QDs can be easily isolated with a magnetic field leading to sample concentration and an interference-free FL signal modulation. The extent of the core-satellite immunocomplexing was directly proportional to influenza virus (H1N1) concentration; this in turn led to stronger plasmonic coupling effect as more MP-MoO3 QDs are brought closer to the gCNQDs by the antibody-antigen binding affinity. Consequently, corresponding linear calibration curves were plotted to elucidate the linear dynamic detection range as shown in Fig. 4C. The LOD for the detection of influenza virus (H1N1) in ultrapure water is 0.25 pg/mL, while the LOD in human serum was evaluated to be 0.9 pg/mL. A comparison of the obtained LODs with other detection systems indicated that this developed system has a comparable sensitivity (Table 1 ) [[48], [49], [50], [51], [52], [53]]. Furthermore, by analyzing the kinetics of the detection cycle of the time-course change in the FL intensity of gCNQDs (Fig. S6 in Supplementary data), it can be observed that the FL enhancement occurred rapidly when the target virus was introduced to the 96 well-plate reaction chamber that allowed rapid fluorescence detection up to 5 min after which no major FL modifications had been observed up to 10 min and above after this optimum point. The detection protocol was expedited for influenza virus (H1N1) detection in a matter of ∼5 min even in clinical samples containing target influenza virus. Hence, this developed detection platform could achieve faster results for urgent diagnostic measures better than or replace commercial rapid diagnostic kits, which requires ∼15 – 20 min, and/or (qRT-PCR), which requires several hours (4–6 h) for detection, such that point-of-care testing could be expected if this detection protocol is fully integrated with portable instrumentation system. Moreover, it is plausible to point out that this detection platform can be tuned appropriately to detect other target viruses.
Fig. 4.
H1N1 detection via enhancement of the FL emission of gCNQDs measured in ultrapure water (A) or human serum (B) and their calibration plots (C). λex = 500 nm.
Table 1.
A comparison of some recent reports/results on influenza virus detection systems.
| Method of detection | Target virus | LOD | Ref. |
|---|---|---|---|
| Fluorescence fiber-optic biosensor | H1N1 | 13.9 pg/mL | [48] |
| Magnetofluoro-immunoassay | H1N1 | 6.07 pg/mL | [49] |
| Ag-S covalent labelling | H1N1 | 0.1 pg/mL | [50] |
| Electrochemical immunosensor | H5N1 | 2.1 pg/mL | [51] |
| Metal-enhanced fluoroimmunoassay | H1N1 | 1 ng/mL | [52] |
| Peroxidase mimic | H1N1 | 10 pg/mL | [53] |
| Magnetoplasmonic fluoroimmunoassay | H1N1 | 0.25 pg/mL (DI water) and 0.9 pg/mL (in serum) | This work |
3.5. Specific recognition of influenza virus (H1N1)
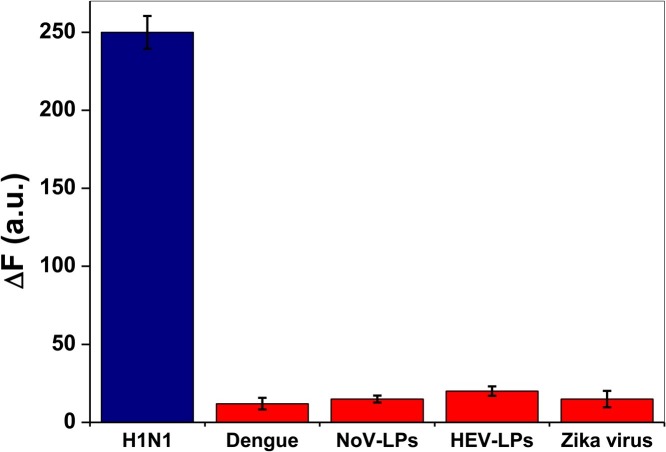
To verify the specificity and selective disposition of the developed immunosensor towards influenza virus (H1N1) as the target virus, some other viruses such as Zika virus, NoV-LPs, HEV-LPs, Dengue were employed to study the potential interferences that may be exhibited by monitoring the response of the fluoroimmunosensors in the presence of ∼ 10 ng/mL other viruses. As shown in Fig. 5 , the specificity of H1N1 is proved by the marked difference in the induced change in the FL intensity of the fluoroimmunosensor signals compared to other non-specific viruses which exhibited very negligibly change in signals. It is pertinent to state here, that the specificity of the fluoroimmunosensor is mainly dependent on the antibodies involved in the immuno-reactions. The specific affinity of the influenza virus to be confined in core-satellite system with the specific Ab-conjugated nanomaterials resulted in the substantial selectivity of the developed biosensor. In addition, to highlight the contributions of gCNQD's surface chemistry to the immunoassay specificity and sensitivity, it is appropriate to stress that hemagglutinin (HA), a major envelope of influenza A virus glycoprotein is composed of HA1 (positively charged), HA2 and signal peptides [54]. Nonetheless, the protein-associated amino acids in the influenza A virus may exhibit a general charge-neutrality at physiological pH [55]. Therefore, electrostatic interaction / repulsion effects are minimized between the influenza A virus and gCNQDs (-18.1 mV) or Ab-gCNQDs (−23.5 mV) (Fig. S3 in Supplementary data). It can therefore be inferred that the influenza A virus' avidity towards the antibody-conjugated gCNQDs / MP-MoO3 QDs resulted in the immunoreactions leading to the specific and sensitive virus detection.
Fig. 5.
The selectivity result of gCNQDs/MP-MoO3 QDs-based fluoroimmunosensor for influenza A/H1N1 detection in the presence of 10 ng/mL of other virus/VLPs and 103 PFU/mL of H3N2 as negative control.
3.6. Detection of clinically isolated influenza virus (H3N2 RNA) and comparison with a commercial test kit
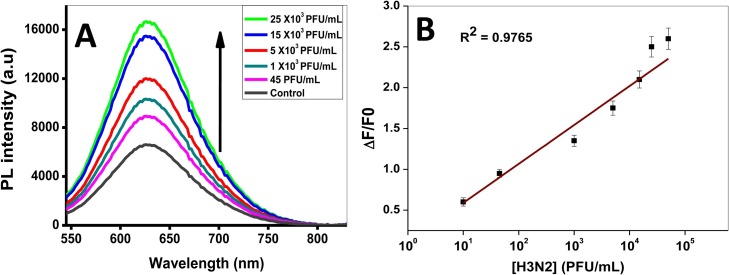
In order to boost the analytical figures of merit of our novel detection system, clinically isolated influenza virus A/Yokohama/110/2009 (H3N2) was tested using the Ab-(H3N2)-conjugated MP-MO3 QDs. The specimen sample containing known PFU/mL of influenza virus (H3N2 RNA) was subjected to biosensing detection within the range of 45 – 25,000 PFU/mL, which triggered the FL enhancement of the probe (Fig. 6 A and B). The LOD was evaluated to be 45 PFU/mL. The performance of developed fluoroimmunoassay was further evaluated against a commercial rapid influenza diagnostic kit (RIDT) – QuikNavi-Flu 2 (Denka Seiken Co. Ltd, Tokyo, Japan). For the RIDT, the concentration of <5000 PFU/mL of the clinically isolated influenza virus A/Yokohama/110/2009 (H3N2) could not be detected (Fig. S7 in Supplementary data). Whereas our developed system is responsive to the virus level up to 45 PFU/mL. This result indicates that our sensing strategy is ∼100 times as sensitive as the commercial RIDT. However, it is pertinent to point that both detection techniques are quite different in principle and the development of a rapid diagnostic kit with our designed system may significantly improve the sensitive detection of influenza virus.
Fig. 6.
(A) FL emission spectra of gCNQDs showing the detection of clinically isolated influenza virus RNA (H3N2) and (B) the corresponding H3N2 detection calibration plot within the linear range. λex = 500 nm.
4. Conclusion
A novel combination of graphitic carbon nitride and molybdenum-based QDs was deployed for the fluoroimmunoassay of influenza virus. The sensitivity achieved herein for influenza virus detection was 0.25 pg/mL in DI water and 0.9 pg/mL in human serum. In a clinical sample, influenza virus A (H3N2) was detected using this assay with LOD of 45 PFU/mL within a linear dynamic detection range of 45 – 25,000 PFU/mL. The assay showed a good sensitivity for the detection of influenza virus in samples with complex matrices owing to the magnetic separation/purification protocol of the assay. This work shows that the nanoparticles combination can be adopted as potential materials for constructing efficient platforms for virus detection. In addition, they are highly competitive and low cost alternatives deployable for the plasmonic-induced and optical detection of infectious viral biomolecules by using the desired antigen-antibody pair to devise a vast pool of biosensors to meet the demand for speedy and responsive assessment assays.
CRediT authorship contribution statement
Ojodomo J. Achadu: Conceptualization, Methodology, Writing - original draft. Kenshin Takemura: Methodology. Indra Memdi Khoris: Investigation. Enoch Y. Park: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors sincerely thank Professor K. Morita of the Institute of Tropical Medicine, Nagasaki University and Professor T. Suzuki of Hamamatsu University School of Medicine for providing the Zika virus and influenza virus A/H3N2, respectively. O.J.A gratefully thanks the Japan Society for the Promotion of Science (JSPS) for a Postdoctoral Fellowship for Research in Japan (Standard) (Grant No. 19F19348).
Biographies
Ojodomo J. Achadu is a JSPS postdoctoral research fellow at Shizuoka University, Japan. He obtained his Ph.D. degree in Chemistry (2017) from Rhodes University, South Africa. His research is focused on biosensors fabrication using functional nanomaterials, optical spectroscopy and materials chemistry. He also has research interests in the photophysics of nanocomposites of carbon-based nanoparticles and macrocyclic organic complexes such as porphyrins and phthalocyanines.
Kenshin Takemura is currently a graduate student in Graduate school of Integrated Science and Technology, Shizuoka University. He received his MS degree in Applied Biological Chemistry in 2018 from Shizuoka University, Japan. He is studying for Ph.D study in Department of Bioscience at Graduate School of Integrated Science and Technology Shizuoka University. His current research interest includes synthesis of functional nanoparticles and biosensor applications. He is also interested in localized surface plasmon resonance of nanocomposites for biosensing application.
Indra Memdi Khoris is currently pursuing his Ph.D study in Department of Bioscience at Graduate School of Integrated Science and Technology Shizuoka University. He received his MS degree in Department of Applied Biological Chemistry, Shizuoka University. His research interest is mainly on the utilization of nanomaterials for virus sensing application.
Enoch Y. Park is currently a professor in Research Institute of Green Science and Technology, Shizuoka University. He received his MS degree in Korea Advanced Institute of Science and Technology in 1982, and Ph.D. degree in a major of Chemical Engineering from the University of Tokyo, Japan in 1990, and worked as assistant professor in the department of chemical engineering Nagoya University for two years. He is currently interested in preparation and engineering of nanobiomaterials such as virus-like particles and detection of infectious viruses. Also, he is interested in the expression of eukaryotic proteins in silkworm larvae and their applicability on biological detection of molecules.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2020.128494.
Contributor Information
Ojodomo J. Achadu, Email: ojodomo.john.achadu@shizuoka.ac.jp.
Kenshin Takemura, Email: takemura.kenshin.16@shizuoka.ac.jp.
Indra Memdi Khoris, Email: indra.memdi.khoris.17@shizuoka.ac.jp.
Enoch Y. Park, Email: park.enoch@shizuoka.ac.jp.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.World Health Organization (WHO), Coronavirus disease 2019 (COVID-19) Situation Report–24, Feb. 13. 2020 (adapted from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports).
- 2.Achadu O.J., Nyokong T. In situ one-pot synthesis of graphitic carbon nitride quantum dots and its 2, 2, 6, 6-tetramethyl (piperidin-1-yl) oxyl derivatives as fluorescent nanoprobes for ascorbic acid. Anal. Chim. Acta. 2017;991:113–126. doi: 10.1016/j.aca.2017.07.047. [DOI] [PubMed] [Google Scholar]
- 3.Benítez-Martínez S., Valcárcel M. Graphene quantum dots in analytical science. TRAC-TREND ANAL Chem. 2015;72:93–113. [Google Scholar]
- 4.Dong Y., Wang Q., Wu H., Chen Y., Lu C.H., Chi Y., Yang H.H. Graphitic carbon nitride materials: sensing, imaging and therapy. Small. 2015;12:5376–5393. doi: 10.1002/smll.201602056. [DOI] [PubMed] [Google Scholar]
- 5.Li H., Shao F.Q., Huang H., Feng J.J., Wang A.J. Eco-friendly and rapid microwave synthesis of green fluorescent graphitic carbon nitride quantum dots for vitro bioimaging. Sens. Actuators B Chem. 2016;226:506–511. [Google Scholar]
- 6.Ngo Y.L.T., Choi W.M., Chung J.S., Hur S.H. Highly biocompatible phenylboronic acid-functionalized graphitic carbon nitride quantum dots for the selective glucose sensor. Sens. Actuators B Chem. 2019;282:36–44. [Google Scholar]
- 7.Zhu S., Song Y., Zhao X., Shao J., Zhang J., Yang B. The photoluminescence mechanism in carbon dots (graphene quantum dots, carbon nanodots, and polymer dots): current state and future perspective. Nano Res. 2015;8:355–381. [Google Scholar]
- 8.Barman S., Sadhukhan M. Facile bulk production of highly blue fluorescent graphitic carbon nitride quantum dots and their application as highly selective and sensitive sensors for the detection of mercuric and iodide ions in aqueous media. J. Mater. Chem. 2012;22:21832–21837. [Google Scholar]
- 9.Pang X., Bian H., Wang W., Liu C., Khan M.S., Wang Q., Qi J., Wei Q., Du B. A bio-chemical application of N-GQDs and g-C3N4 QDs sensitized TiO2 nanopillars for the quantitative detection of pcDNA3-HBV. Biosens. Bioelectron. 2017;91:456–464. doi: 10.1016/j.bios.2016.12.059. [DOI] [PubMed] [Google Scholar]
- 10.Sun B., Dong J., Cui L., Feng T., Zhu J., Liu X., Ai S. A dual signal-on photoelectrochemical immunosensor for sensitively detecting target avian viruses based on AuNPs/g-C3N4 coupling with CdTe quantum dots and in situ enzymatic generation of electron donor. Biosens. Bioelectron. 2019;124–125:1–7. doi: 10.1016/j.bios.2018.09.100. [DOI] [PubMed] [Google Scholar]
- 11.Xiong M.Y., Rong Q.M., Meng H.M., Zhang X.B. Two-dimensional graphitic carbon nitride nanosheets for biosensing applications. Biosens. Bioelectron. 2017;89:212–223. doi: 10.1016/j.bios.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Rasheed P.A., Radhakrishnan T., Nambiar S.R., Thomas R.T., Sandhyarani N. Graphitic carbon nitride as immobilization platform for ssDNA in a genosensor. Sens. Actuators B Chem. 2017;250:162. [Google Scholar]
- 13.Holá K., Sudolská M., Kalytchuk S., Nachtigallová D., Rogach A.L., Otyepka M., Zbořil R. Graphitic nitrogen triggers red fluorescence in carbon dots. ACS Nano. 2017;12:12402–12410. doi: 10.1021/acsnano.7b06399. [DOI] [PubMed] [Google Scholar]
- 14.Li C., Yang X., Yang B., Yan Y., Qian Y. Synthesis and characterization of nitrogen-rich graphitic carbon nitride. Mater. Chem. Phys. 2007;103:427–432. [Google Scholar]
- 15.Zhu X., Kou F., Xu H., Han Y., Yang G., Huang X., Chen W., Chi Y., Lin Z. Label-free ochratoxin A electrochemical aptasensor based on target-induced non-covalent assembly of peroxidase-like graphitic carbon nitride nanosheets. Sens. Actuators B Chem. 2018;270:263–269. [Google Scholar]
- 16.Han T., Li X., Li Y., Cao W., Wu D., Du B., Wei Q. Gold nanoparticles enhanced electrochemiluminescence of graphite-like carbon nitride for the detection of Nuclear Matrix Protein 22. Sens. Actuators B Chem. 2014;205:176–183. [Google Scholar]
- 17.Wang H., Zhang Q., Yin H., Wang M., Jiang W., Ai S. Photoelectrochemical immunosensor for methylated RNA detection based on g-C3N4/CdS quantum dots heterojunction and Phos-tag-biotin. Biosens. Bioelectron. 2017;95:124–130. doi: 10.1016/j.bios.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Achadu O.J., Revaprasadu N. Microwave-assisted synthesis of thymine-functionalized graphitic carbon nitride quantum dots as fluorescent nanoprobe for mercury (II) Microchim. Acta. 2018;185:461–469. doi: 10.1007/s00604-018-3004-2. [DOI] [PubMed] [Google Scholar]
- 19.Achadu O.J., Revaprasadu N. Tannic acid-derivatized graphitic carbon nitride quantum dots as an “on-off-on” fluorescent nanoprobe for ascorbic acid via copper(II) mediation. Microchim. Acta. 2019;186:87–96. doi: 10.1007/s00604-018-3203-x. [DOI] [PubMed] [Google Scholar]
- 20.Tao H., Hu T., Yan J., Di J. A comparative study of different reagentless plasmon sensors based on Ag–Au alloy nanoparticles for detection of Hg. Sens. Actuators B Chem. 2015;208:43–49. [Google Scholar]
- 21.Liu C., Meng F., Zheng W., Xue T., Jin Z., Wang Z., Cui X. Plasmonic ZnO nanorods/Au substrates for protein microarrays with high sensitivity and broad dynamic range. Sens. Actuators B Chem. 2016;228:231–236. [Google Scholar]
- 22.Lee S.H., Nishi H., Tatsuma T. Tunable plasmon resonance of molybdenum oxide nanoparticles synthesized in non-aqueous media. Chem. Commun. 2017;53:12680–12683. doi: 10.1039/c7cc08090e. [DOI] [PubMed] [Google Scholar]
- 23.Lu X., Wang R., Yang F., Iao W., Liu W., Hao L., He X. Preparation of MoO3 QDs through combining intercalation and thermal exfoliation. J. Mater. Chem. C. 2016;4:6720–6726. [Google Scholar]
- 24.Shi J., Kuwahara Y., Wen M., Navlani-Garcia M., Mori K., An T., Yamashita H. Room‐temperature and aqueous‐phase synthesis of plasmonic molybdenum oxide nanoparticles for visible‐light‐Enhanced hydrogen generation. Chem. Asian J. 2016;11:2377–2381. doi: 10.1002/asia.201600771. [DOI] [PubMed] [Google Scholar]
- 25.Xiao S.J., Zhao X.J., Hu P.P., Chu J.Z., Huang C.Z., Zhang L. Highly photoluminescent molybdenum oxide quantum dots: one-pot synthesis and application in 2,4,6-Trinitrotoluene determination. ACS Appl. Mater. Interfaces. 2016;8:8184–8191. doi: 10.1021/acsami.5b11316. [DOI] [PubMed] [Google Scholar]
- 26.Yin H., Kuwahara Y., Mori K., Cheng H., Wen M., Huo Y., Yamashita H. Localized surface plasmon resonances in plasmonic molybdenum tungsten oxide hybrid for visible-light-enhanced catalytic reaction. J. Phys. Chem. C. 2017;42:23531–23540. [Google Scholar]
- 27.Zhang J., Pan Y., Chen Y., Lu H. Plasmonic molybdenum trioxide quantum dots with noble metal-comparable surface enhanced Raman scattering. J. Mater. Chem. C. 2018;6:2216–2220. [Google Scholar]
- 28.Zu H., Guo Y., Yang H., Huang D., Liu Z., Liu Y., Hu C. Rapid room-temperature preparation of MoO3−x quantum dots by ultraviolet irradiation for photothermal treatment and glucose detection. New J. Chem. 2018;42:18533–18540. [Google Scholar]
- 29.Lee J., Kim H., Algar R. Thiol-ligand-Catalyzed quenching and etching in mixtures of colloidal quantum dots and silver nanoparticles. J. Phys. Chem. C. 2017;121:28566–28575. [Google Scholar]
- 30.Vaishnav J.K., Mukherjee T.K. Long-range resonance coupling-induced surface energy transfer from CdTe quantum dot to plasmonic nanoparticle. J. Phys. Chem. C. 2018;122:28324–28336. [Google Scholar]
- 31.Adegoke O., Morita M., Kato T., Ito M., Suzuki T., Park E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017;94:513–522. doi: 10.1016/j.bios.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 32.Lee J., Ahmed S.R., Oh S., Kim J., Suzuki T., Parmar K., Park S., Lee J., Park E.Y. A plasmon-assisted fluoro-immunoassay using gold nanoparticle-decorated carbon nanotubes for monitoring the influenza virus. Biosens. Bioelectron. 2015;64:311–317. doi: 10.1016/j.bios.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 33.Takemura K., Adegoke O., Naoto T., Kato T., Li T.C., Kitamoto N., Tanaka T., Suzuki T., Park E.Y. Versatility of a localized surface plasmon resonance-based gold nanoparticle-alloyed quantum dot nanobiosensor for immunofluorescence detection of viruses. Biosens. Bioelectron. 2017;89:998–1005. doi: 10.1016/j.bios.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 34.Takemura K., Lee J., Suzuki T., Hara T., Abe F., Park E.Y. Ultrasensitive detection of norovirus using a magnetofluoroimmunoassay based on synergic properties of gold/magnetic nanoparticle hybrid nanocomposites and quantum dots. Sens. Actuators B Chem. 2019;296:126672–126680. [Google Scholar]
- 35.Nasrin F., Chowdhury A.D., Takemura K., Lee J., Adegoke O., Deo V.K., Abe F., Suzuki T., Park E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018;122:16–24. doi: 10.1016/j.bios.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed S.R., Takemura K., Li T.C., Kitamoto N., Tanaka T., Suzuki T., Park E.Y. Size-controlled preparation of peroxidase-like graphene-gold nanoparticle hybrids for the visible detection of norovirus-like particles. Biosens. Bioelectron. 2017;87:558–565. doi: 10.1016/j.bios.2016.08.101. [DOI] [PubMed] [Google Scholar]
- 37.Li T.C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M., Uchida T., Takeda N., Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarei M., Ahmadzadeh H., Goharshadi E.K., Farzaneh A. Graphitic carbon nitride embedded hydrogels for enhanced gel electrophoresis. Anal. Chim. Acta. 2015;887:245–252. doi: 10.1016/j.aca.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 39.Lima K.M.G., Raimundo I.M., Jr., Pimentel M.F. Improving the detection limits of near infrared spectroscopy in the determination of aromatic hydrocarbons in water employing a silicone sensing phase. Sens. Actuators B Chem. 2007;125:229–233. [Google Scholar]
- 40.Loo A.H., Sofer Z., Bouša D., Ulbrich P., Bonanni A., Pumera M. Carboxylic carbon quantum dots as a fluorescent sensing platform for DNA detection. ACS Appl. Mater. Interfaces. 2016;8:1951–1957. doi: 10.1021/acsami.5b10160. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., Mintz K.J., Sharma S.K., Leblanc R.M. Carbon dots: diverse preparation, application, and perspective in surface chemistry. Langmuir. 2019;35:9115–9132. doi: 10.1021/acs.langmuir.9b00595. [DOI] [PubMed] [Google Scholar]
- 42.Ding H., Yu S.B., Wei J.S., Xiong H.M. Full-color light-emitting carbon dots with a surface-state-controlled luminescence mechanism. ACS Nano. 2016;10:484–491. doi: 10.1021/acsnano.5b05406. [DOI] [PubMed] [Google Scholar]
- 43.Yuan F., Wang Z., Li X., Li Y., Tan Z., Fan L., Yang S. Bright multicolor band-gap fluorescent carbon quantum dots for electroluminescent light‐emitting diodes. Adv. Mater. 2017;29:1604436–1604442. doi: 10.1002/adma.201604436. [DOI] [PubMed] [Google Scholar]
- 44.Gu S.Y., Hsieh C.T., Gandomi Y.A., Chang J.K., Li J., Li J.L., Zhang H.A., Guo Q., Lau K.C., Pandey R. Microwave growth and tunable photoluminescence of nitrogen-doped graphene and carbon nitride quantum dot. J. Mater. Chem. C. 2019;7:5468–5476. [Google Scholar]
- 45.Wang J., Cao S., Ding Y., Ma F., Lu W., Sun M. Theoretical investigations of optical origins of fluorescent graphene quantum dots. Sci. Rep. 2016;6:24850–24855. doi: 10.1038/srep24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neiva E.G.C., Bergamini M.F., Oliveira M.M., Marcolino L.H., Zarbin A.J.G. PVP-capped nickel nanoparticles: synthesis, characterization and utilization as a glycerol electrosensor. Sens. Actuators B Chem. 2014;196:574–581. doi: 10.1016/j.snb.2014.02.041. [DOI] [Google Scholar]
- 47.Plácido A., Pereira C., Guedes A., Barroso M.F., Miranda-Castro R., Santos-Álvarez N.D., Delerue-Matos C. Electrochemical genoassays on gold-coated magnetic nanoparticles to quantify genetically modified organisms (GMOs) in food and feed as GMO percentage. Biosens. Bioelectron. 2018;110:147–154. doi: 10.1016/j.bios.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 48.Chang Y.F., Wang S.F., Huang J.C., Su L.C., Yao L., Li Y.C., Wu S.C., Chen Y.M., Hsieh J.P., Chou C. Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2010;26:1068–1073. doi: 10.1016/j.bios.2010.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J., Takemura K., Park E.Y. Plasmonic/magnetic graphene-based magnetofluoro-immunosensing platform for virus detection. Sens. Actuators B Chem. 2018;276:254–261. [Google Scholar]
- 50.Li Y., Hong M., Qiu B., Lin Z., Chen Y., Cai Z., Chen G. Highly sensitive fluorescent immunosensor for detection of influenza virus based on Ag autocatalysis. Biosens. Bioelectron. 2014;54:358–364. doi: 10.1016/j.bios.2013.10.045. [DOI] [PubMed] [Google Scholar]
- 51.Jarocka U., Sawicka R., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecki J., Radecka H. Electrochemical immunosensor for detection of antibodies against influenza A virus H5N1 in hen serum. Biosens. Bioelectron. 2014;55:301–306. doi: 10.1016/j.bios.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 52.Ahmed S.R., Hossain M.A., Park J.Y., Kim S.H., Lee D., Suzuki T., Lee J., Park E.Y. Metal enhanced fluorescence on nanoporous gold leaf-based assay platform for virus detection. 2014. Biosens. Bioelectron. 2014;58:33–39. doi: 10.1016/j.bios.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed S.R., Kim J., Suzuki T., Lee J., Park E.Y. Detection of influenza virus using peroxidase-mimic of gold nanoparticles. Biotechnol. Bioeng. 2016;113:2298–2303. doi: 10.1002/bit.25982. [DOI] [PubMed] [Google Scholar]
- 54.Arinaminpathy N., Grenfell B. Dynamics of glycoprotein charge in the evolutionary history of human influenza. PLoS One. 2010;5:1–7. doi: 10.1371/journal.pone.0015674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi Y., Suzuki Y. Compensatory evolution of net-charge in influenza a virus hemagglutinin. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.