Abstract
A previously unknown coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been shown to cause coronavirus disease 2019 (COVID-19) pandemic. The first case of COVID-19 in Turkey has been declared in March 11th, 2020 and from there on, more than 150,000 people in the country have been diagnosed with the disease. In this study, 62 viral sequences from Turkey, which have been uploaded to GISAID database, were analyzed by means of their nucleotide substitutions in comparison to the reference SARS-CoV-2 genome from Wuhan. Our results indicate that the viral isolates from Turkey harbor some common mutations with the viral strains from Europe, Oceania, North America and Asia. When the mutations were evaluated, C3037T, C14408T and A23403G were found to be the most common nucleotide substitutions among the viral isolates in Turkey, which are mostly seen as linked mutations and are part of a haplotype observed high in Europe.
Keywords: SARS-CoV-2, evolution, mutation, COVID-19
1. Introduction
Coronaviruses (CoV) are enveloped positive-stranded RNA viruses that belong to the family Coronaviridae and are divided into 4 genera which are alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV. Similar to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 is a beta-coronavirus (Gorbalenya et al., 2020). It encodes several structural proteins including envelope (E), membrane (M), nucleocapsid (N) and spike (S) proteins as well as non structural ones (Gorbalenya et al., 2020). The virus was shown to have gone through certain mutations both in its structural and non structural proteins within several months while spreading throughout the world (Pachetti et al., 2020; Wang et al., 2020).
Starting from December 2019, SARS-CoV-2 led to a worldwide COVID-19 pandemic, which caused more than 3 million cases along with more than 250,000 deaths within 5 months1. The first case of COVID-19 in Turkey was announced in March 11th and as of May 24th, the number of positive cases and deaths reached to 156,827 and 4,340, respectively2. A total of 63 sequences from SARS-CoV-2 isolates of Turkey were uploaded to global initiative on sharing all influenza data (GISAID) database between the dates March 25th and May 22nd3.
The aim of this study is to reveal the most common mutations of SARS-CoV-2 viral isolates from Turkey in comparison to the reference sequence from China (NC_045512.1). Our results revealed that some of the viral mutations are present in more than 60% of the isolates. Although further analysis and characterizations are needed, the data in this study may contribute to understanding the molecular evolution of SARS-CoV-2 in Turkey.
2. Materials and methods
2.1. Dataset construction
All the SARS-CoV-2 whole genome sequences have been downloaded from GISAID database3 (Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017). The whole genome sequence dataset was constructed as including 63 viral sequences from Turkish patients that were submitted to the database between March 25th and May22nd, 2020 (Supplementary Table 1) and the reference SARS-CoV-2 sequence isolated in Wuhan which was downloaded from GeneBank (NC_045512.1)4. One of the sequences (EPI_ISL_435057) was excluded from the analysis due to harboring extreme number of unique mutations, which can result from sequencing errors.
Table 1.
Common nucleotide substitutions in 62 SARS-CoV-2 viral genomes fromTurkey (submitted to GISAID between March 25th and May 22nd 2020) compared to the SARS-CoV-2 NCBI reference genome NC 045512.1. The viral gene and gene products were identified according to the reference sequence information from GeneBank4. The nucleotide sequences were indicated starting from the 5’ UTR, while the corresponding amino acid changes were mentioned separately for each protein coding region specific for the corresponding protein.
| Nucleotide substitution at the given position | Corresponding viral gene | Corresponding viral gene product | Amino acid change within the corresponding protein (if exists) | Mutation type | Number of samples seen (among 62 samples) | Percentage among 62 samples |
|---|---|---|---|---|---|---|
| C3037T | ORF1a | Nsp3 | 106 (F) | Silent | 38 | 61% |
| C14408T | ORF1b | RNA-dependent RNA polymerase | P323L | Missense | 38 | 61% |
| A23403G | S | Spike glycoprotein | D614G | Missense | 38 | 61% |
| G25563T | ORF3a | ORF3a protein | Q57H | Missense | 25 | 40% |
| G11083T | ORF1a | Nsp6 | L37F | Silent | 24 | 38% |
| C18877T | ORF1b | 3’ to 5’ exonuclease | 280 (L) | Silent | 22 | 35% |
| G29742T | 3’ UTR | 22 | 35% | |||
| G1397A | ORF1a | Nsp2 | V198I | Missense | 21 | 33% |
| T28688C | N | Nucleocapsid phosphoprotein | 139(L) | Silent | 21 | 33% |
| C241T | 5’ UTR | 20 | 32% | |||
| C26735T | M | Membrane glycoprotein | 71(Y) | Silent | 13 | 20% |
| C26549T | M | Membrane glycoprotein | 9(T) | Silent | 12 | 19% |
| C884T | ORF1a | Nsp2 | R27C | Missense | 11 | 17% |
| G8653T | ORF1a | Nsp4 | M33I | Missense | 11 | 17% |
| G28881A | N | Nucleocapsid phosphoprotein | R203K | Missense | 9 | 14% |
| G28882A | ||||||
| G28883C | N | Nucleocapsid phosphoprotein | G204R | Missense | 9 | 14% |
| C228T | 5’ UTR | 8 | 13% | |||
| A9514G | ORF1a | Nsp4 | 320(L) | Silent | 7 | 11% |
| C22444T | S | Spike glycoprotein | 294(D) | Silent | 7 | 11% |
| G26720C | M | Membrane glycoprotein | 66(V) | Silent | 7 | 11% |
| C28854T | N | Nucleocapsid phosphoprotein | S194L | Missense | 7 | 11% |
| C5736T | ORF1a | Nsp3 | A1006V | Missense | 6 | 10% |
| G9479T | ORF1a | Nsp4 | G309C | Missense | 6 | 10% |
| T28835C | N | Nucleocapsid phosphoprotein | S188P | Missense | 6 | 10% |
| C7765T | ORF1a | Nsp3 | 1682(S) | Silent | 5 | 8% |
| C17690T | ORF1b | Helicase | S485L | Missense | 5 | 8% |
| T26551C | M | Membrane glycoprotein | V10A | Missense | 5 | 8% |
2.2. Nucleotide substitution analysis
SARS-CoV-2 isolate sequences from Turkey were compared to the reference SARS-CoV-2 sequence (NC_045512.1), by means of nucleotide substitutions. The constructed dataset was MAFFT5 aligned and manually edited using the AliView program to verify that the sequences were in frame. The nucleotide numbers were indicated starting from the 5’ UTR of the viral sequence, while amino acid positions were indicated separately for each corresponding protein coding region4. The positions of the nucleotides and amino acids were further confirmed from GeneBank reference sequences (NC_045512.1)4. The nonconserved nucleotide positions were determined and the nucleotide substitutions were evaluated for their effects on amino acid changes by using the AliView and MEGA software.
3. Results
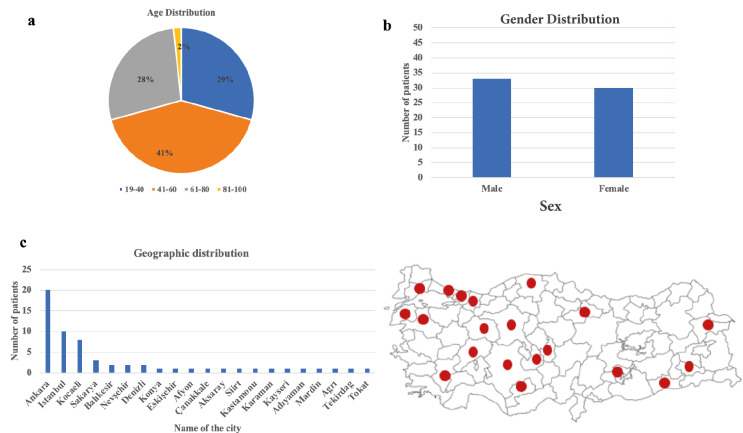
The age interval of the patients from Turkey, from whom the viral isolates were taken, was between 19 and 82. 41% of the patients were between the age range of 41–60, while 29% was between 19 and 40 and 28% was between 61 and 80 (Figure 1a) and the sex distribution was approximately equal (Figure 1b). Most of the uploaded samples seem to come from middle-west side of the country (Figure 1c).
Figure 1.
a)The age, b) Sex and c)Geographic distribution of the 62 patients from whom the viral isolates were taken.
When the SARS-CoV-2 strains from Turkey, compared to the reference viral sequence (NC 045512.1), some missense and silent mutations were identified. In more than 40% of the viral isolates, one or more of nucleotide substitutions of C3037T, C14408T ,A23403G, and G25563T were observed, which are present in the coding regions for the Nsp3, RNA-dependent RNA polymerase, spike glycoprotein and ORF3a protein, respectively (Table 1). Among these mostly seen substitutions, the ones seen in Nsp3, RNA-dependent RNA polymerase and spike glycoprotein were present in 61% (38/62) of the isolates from Turkey. Other viral genome regions where the nucleotide substitutions were observed in 5 or more samples out of 62 include, Nsp6 (24/62), 3’ to 5’ exonuclease (22/62), Nsp2 (22/62), nucleocapsid protein (21/62), membrane glycoprotein (13/62), Nsp4 (11/62), Nsp3 (6/62) and Helicase (5/62) (Table 1).
Each of the 62 viral isolates were also evaluated by means of the mutations they harbor and 24 viral isolates found to have unique mutations in addition to the common mutations they harbor, that are not seen in other isolates from Turkey (Table 2; Supplementary Table 2).
Table 2.
Nucleotide substitutions present only in a single isolate among the analyzed viral isolates from Turkey.
| GISAID ID | Nucleotide substitutions present only in the corresponding isolate |
|---|---|
| EPI_ISL_424366 | G23876A, C29563T |
| EPI_ISL_427391 | C2997T |
| EPI_ISL_428368 | C12809T |
| EPI_ISL_428717 | C21304A, G21305A, C28054T |
| EPI_ISL_428718 | C8782T, G14122T, G28878A |
| EPI_ISL_428720 | G12248T, T23559A |
| EPI_ISL_428713 | C4524T |
| EPI_ISL429870 | C19170T, C25275T |
| EPI_ISL_429873 | C1437T |
| EPI_ISL_429864 | G944A |
| EPI_ISL429865 | C7834T, C26340T |
| EPI_ISL_429868 | C11074T |
| EPI_ISL_437306 | C8683A |
| EPI_ISL_437307 | T6202A, C8964T, C10202T, C16247T, C24865T |
| EPI_ISL_437308 | C15240T |
| EPI_ISL_437309 | C16616T, A23734T |
| EPI_ISL_437317 | G22468T, G25314T, T28144C |
| EPI_ISL_437318 | C5477T, C6402T |
| EPI_ISL_437319 | G19285A |
| EPI_ISL_437328 | C1825T |
| EPI_ISL_437330 | C5826A |
| EPI_ISL_437331 | C12700T |
| EPI_ISL_437333 | T15102C |
| EPI_ISL_437335 | A27354G |
Most of the viral isolates found to have different nucleotide substitution combinations. However, the same nucleotide substitutions were observed for the samples EPI_ISL42874 and EPI_ISL_429871. Simlarly, EPI_ISL437411 and EPI_ISL437413 were also found to have the same nucleotide substitution combination among them (Supplementary Table 2). The analyzed viral isolates were found to harbor 4 to12 mutations per isolate compared to the reference sequence (Supplementary Table 2). C > T mutations also observed to predominate among the analyzed viral isolates.
4. Discussion
The first COVID-19 case in Turkey was declared in March 11th, almost after two and a half months from the first case declaration in China. When the nucleotide substitutions for the SARS-CoV-2 isolates in Turkey were analyzed compared to the reference genome of the virus from China, it was seen that during this time period, the virus had undergone several nucleotide substitutions, including silent and missense mutations.
When we consider the mutations in ORF1ab, nucleotide substitutions of C884T, G1397A, C3037T, G8653T, G11083T, C14408T, and C18877T were seen in more than 15% (11/62) of the samples. C14408T mutation within the RNA-dependent RNA polymerase encoding region of ORF1b, which is a missense mutation that leads to an amino acid change from proline to leucine at position 323 (P323L) in RNA polymerase protein, was amongst the most commonly seenmutations [61% (38/62)] in isolates from Turkey. Both amino acids seem to have similar isoelectric points and this mutation is mostly seen in isolates from Europe, followed by North America (Pachetti et al., 2020). The mutation was found to be present in European isolates after February 20th, 2020 and thought to be associated with increased number of point mutations compared to isolates from Asia, which proposed to be somehow due to the presence of RNA polymerase within the proofreading machinery of the virus (Pachetti et al., 2020). A recent study indicates that SARS-CoV-2 genomes which harbor C14408T mutation, are more likely to have mutations in the membrane (M) and envelope (E) proteins (Eskier et al., 2020). Furthermore, recently revealed structure of the replicating RNA polymerase of SARS-CoV-2 may further help to understanding of the effect of certain mutations within this protein (Hillen et al., 2020).
G11083T, corresponding to the amino acid substitution L37F within Nsp6 protein, was present in 38% of the samples (24/62) and this mutation was previously seen in SARS-CoV-2 sequences analyzed from all over the world (Benvenuto et al., 2020; Wang et al., 2020). In the study of Wang et al. (2020), C8782T substitution, which is also a silent mutation, was present in 28 out of 95 samples, although this mutation was only present in 2 samples (EPI_ISL_428718 and EPI_ISL_437317) in our study, which indicates that this mutation is not as common as in Europe for the viral isolates in Turkey and may be related with isolates from other countries. Both C8782T and G11083T mutations were found to be mostly present in Oceania isolates, where followed by North America and Europe subsequently (Pachetti et al., 2020). G1397A substitution in Nsp2 encoding region of ORF1a, which was present in 33% (21/62) of the isolates, was mainly seen in viral isolates from Oceania, however was also present in minor amounts in isolates from Asia and North America (Pachetti et al., 2020). This substitution leads to an amino acid change from valine to isoleucine at the position 198 (V198I) within Nsp2 protein, where both amino acids have the same isoelectric points.
A23403G mutation in the spike glycoprotein coding region was also amongst the mostly seen mutations in viral isolates from Turkey (61%). Spike glycoprotein functions to bind target receptor and facilitate membrane fusion and viral entry (Ou et al., 2020). This protein has 2 subunits, S1 and S2, where the former mediates attachment and the later mediates membrane fusion. A23403G substitution was found to be present in isolates from Europe and leads to an amino acid change from aspartate to glycine at position 614 (D614G) within the spike glycoprotein, where these amino acids differ by means of their isoelectric points (Pachetti et al., 2020). Another mutation found within the spike protein encoding region was C22444T, which is a silent mutation and seen in 7 out of 62 isolates (Table 1).
Similar to SARS-CoV, receptor binding domain (RBD) within the spike glycoprotein of SARS-CoV-2 seems to play a major role in viral infection by acting as an interaction point with target receptors on the host cell surface (Raj et al., 2013). In a recent study which performed multivariate generalized linear model (GLM) analysis with outpatient and hospitalized patients in the Sheffield Teaching Hospitals NHS Foundation Trust as the outcome revealed that patients carrying G614 mutation had higher viral loads compared to D614, although D614G status did not significantly affect the hospitalization status (Korber et al., 2020). It also seems that the viral isolates which carry D614G mutation increases in number across the world and this mutation was proposed to have effect on the viral infectivity either due to its presence on the spike protein promoter surface region which might affect hydrogen bonding properties with neighbouring promoter regions or due to be in a site surrounded by antibody-dependent enhancement targets, where antibody binding may lead to a confirmational change that might increase the ACE2 interaction. Both mechanisms were proposed to play role in a more transmissible form of the virus (Korber et al., 2020). On the other hand, another study perfomed on 15,000 SARS-CoV-2 genomes indicated that the recurrent mutations do not increase transmissibility (Dorp et al., 2020). Therefore, the effect of D614G mutation on the transmission of the virus is still a debate.
Five out of 27 commonly seen mutations in viral isolates from Turkey were within the nucleocapsid phosphoprotein. One interesting finding was the presence of 2 subsequent missense mutations that are seen as a cluster in 9 out of 62 samples (14 %). These mutations were due to nucleotide substitutions in 3 nucleotides in order where 2 of them (G28881A and G28882A) results in arginine to lysine (R203K) substitution and the third one (G28883C) results in glycine to arginine (G204R) in the nucleocapsid phosphoprotein, where both substituted amino acids differ from their original amino acids in means of their isoelectric points (Pachetti et al., 2020) (Table 1). G28881A, along with A23403G substitution in spike glycoprotein, seems to have occurred after February 16th, 2020 in Europe (Pachetti et al., 2020). C28854T substitution in the nucleocapsid protein coding region, which leads to an amino acid change (S194L), was another missense mutation that also was seen in 6/95 samples in a previous study where 95 sequences from different countries were evaluated (Wang et al., 2020). These findings support the presence of the mutation in several strains all over the world including some isolates from Turkey. In addition to S protein, nucleocapsid protein was also proposed to be important in COVID-19 infectivity (Goh et al, 2020). Further studies are needed to clarify if the missense mutations within this region can be important in the infection strategy of the SARS-CoV-2 virus or not.
G1397A, T28688C and G29742T substitutions were said to belong to a monophyletic group which is defined by the presence of these 3 mutations and were found to present in patients who were traveled to or are residents in Iran (Eden J et al., 2020) as well as in Australian and New Zealand isolates. Twenty-one viral isolates from Turkey harbor those 3 mutations together (EPI_ISL_417413, EPI_ISL_424366, EPI_ISL_428722, EPI_ISL_428713, EPI_ISL_429872, EPI_ISL_429865, EPI_ISL_429868, EPI_ISL_437319, EPI_ISL_437324, EPI_ISL_437325, EPI_ISL_437326, EPI_ISL_437327, EPI_ISL_437332, EPI_ISL_437306, EPI_ISL_437307, EPI_ISL_437312, EPI_ISL_437314, EPI_ISL_437320, EPI_ISL_437321, EPI_ISL_437322, EPI_ISL_437323, EPI_ISL_437334). The travel history to Iran were only mentioned for 6 samples in GISAID (EPI_ISL_437319, EPI_ISL_437324, EPI_ISL_437325, EPI_ISL_437326, EPI_ISL_437327, EPI_ISL_437332), where 4 of them (EPI_ISL_437319, EPI_ISL_437324, EPI_ISL_437325,EPI_ISL_437327) are known to harbor this monophyletic group. However, other 2 samples do not harbor any of these mutations, although they have travel history to Iran. Identification of detailed epidemiological data of these samples can be important to identify if these patients somehow had contact in relation to any of these countries.
A phylogenetic network analysis of 160 SARS-CoV-2 genomes, identified 3 central variants of the virus (named as A, B and C), compared to the bat coronavirus (Forster et al., 2020). These variants differ from each other by amino acid substitutions. Node A has 2 subclusters where there is T or C in nucleotide position 29095. B-type variant have T8782C nonsynonymous and C28144T synonymous (Leu to Ser) substitution in addition to A-type and C-type variant have G26144T synonymous mutation (Gly to Val) in addition to B-type substitutions. A- and C-types are said to present mainly outside of East Asia, where B-type is said to be present mainly in East Asia. The isolates from Turkey analyzed in this study mainly harbor cytosine in nucleotide position 29095. Another study, which analyzed 622 complete SARS-CoV-2 genomes by an unrooted maximum likelihood tree divided the viral genomes into 3 clusters, which was mainly similar to the 3 viral variants identified in 162 SARS-CoV-2 genomes (Forster et al., 2020), and performed linkage analysis between the mutations seen within these clusters (Bai et al., 2020). According to the linkage analysis, C241T, C3037T, C14408T and A23403G in Cluster 3 were in complete linkage and the TTTG haplotype was high in Europe and correlated with the death rate. In the viral isolates analyzed in this study, C3037T, C14408T and A23403G, which were the most common mutations (61%), exist together. In 11 out of 62 samples, C241T was also observed to be present together with C3037T, C14408T and A23403G. The reason of not observing C241T in linkage with other mutations with the same percentage can be due to the absence of the first 265 nucleotides in 25 of the uploaded sequences to GISAID. However, there are also isolates that harbor either C241T and not the other 3 mutations or vice versa. This haplotype was proposed to be related with the high death rates in Europe (Bai et al., 2020). Analyzing the course of the COVID-19 disease in patients from whom the viral isolates were taken can give further information about the relatedness of this haplotype with the death rates in Turkey.
Apart from the common mutations, when we consider mutations seen in a single sample among analyzed isolates, some are not mentioned previously in the literature. C8782T was previously seen in more than 10 isolates in Guangdong province of China (Lu et al., 2020) and proposed to be clade specific in a study performed on 313 SARS-CoV-2 genomes (Li, Li, Cui, and Wu, 2020). G28878A, which is present in the same isolate with C8782T, was observed in isolates from Australia and USA (Li et al., 2020). Some of the observed mutations can be either unique to corresponding isolate or can also be a result of homoplasy or sequencing artefact since in an ongoing study, some sites within the viral genome are suspected to be homoplasic substitutions or sequencing artefacts6. G11083T is the most common one among such sites across different countries and sequencing technologies, which might be an indicator of this position being either a site for frequent mutation or an artefact. However, 38% of the samples (24/62) analyzed in this study harbor this mutation, although being sequenced by different technologies, which is consistent with this site being a site for frequent mutation.
Some homoplasic sites were found to bespecific to certain sequencing technologies, such as the nucleotide position 11074. Nucleotide 3037 and 11074 were reported to be either artefacts or hypermutable low-fitness sites. However, 3037 was found to have a linkage with 3 other mutations and mainly observed in Europe. It is also among the mostly seen mutations along with 14408 and 23403 in the viral isolates analyzed. Therefore, it can be 1 of the hypermutable sites within the viral genome. Apart from 11074 and 3037, detailed analysis of the identified unique mutations by means of possible sequencing artefacts and homoplasic sites can reveal more information about them. Therefore, considering the possible sequencing artefacts while analyzing the sequences for substitution can be important (Korber et al., 2020).
Recombination is known to take place in evolution of coronaviruses and some breakpoints for recombination in SARS-CoV-2 was also reported (Korber et al., 2020; Rehman et al., 2020). Therefore, apart from single nucleotide mutations, identification of possible recombinational events can be important in vaccine development strategies.
In our dataset, 2 samples found to harbor quite lots of mutations, which both are sequenced with the same sequencing technology. Therefore, only the common mutations with other isolates in one of them were considered and the other one was excluded totally since the mutations it harbors were quite extreme. The number of extreme mutations might be due to the use of separate sequencing technology in these strains compared to the other isolates that are analyzed in this study.
The nucleotide substitutions showed that viral isolates from Turkey are genetically close to the ones from Europe, Middle East, North America and Asia. C3037T, C14408T and A23403G substitutions, which are present in Nsp3, RNA-dependent RNA polymerase and spike encoding regions respectively, were found to be the mostly seen mutations in Turkey SARS-CoV-2 isolates. Considering the missense mutations encountered in these isolates, further studies are needed how the identified amino acid changes affect the structure of the related proteins as well as the infectivity and spread of the virus. Also, the silent mutations within SARS-CoV-2 genome can be followed up to determine if any further missense mutations will take place within these regions, which may be helpful to understand the evolutionary strategy of the virus as it continues to evolve during its spread through the world.
Acknowledgments/Disclaimers
The authors acknowledge to all the researchers in originating and submitting labs who have shared the SARS-CoV-2 genome data on GISAID ((http://www.gisaid.org/). The extended acknowledgement can be found as a supplementary file. No funding was used to conduct this research.
Supporting Information
General information taken from GISAID about the SARS-CoV-2 sequences used in this study. (NA: Not available)
| GISAID ID | Location | Sex | Age | Collection date | Submission date | Travel history/other | Sequencing technology |
|---|---|---|---|---|---|---|---|
| EPI_ISL_429873 | Kocaeli | Male | 71 | 2020-03-23 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429872 | Kocaeli | Female | 50 | 2020-03-25 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429871 | Ankara | Male | 77 | 2020-03-23 | 2020-04-24 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429870 | Sakarya | Female | 57 | 2020-03-22 | 2020-04024 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429869 | Konya | Female | 59 | 2020-03-17 | 2020-04-24 | Patient travelled to Saudi Arabia | IlluminaMiseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429868 | Eskişehir | Female | 79 | 2020-03-17 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429867 | Balıkesir | Female | 72 | 2020-03-17 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429866 | Afyon | Female | 52 | 2020-03-16 | 2020-04-24 | Patient travelled to Saudi Arabia | IlluminaMiseq Assembly: Burrows-Wheeler Aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429865 | Çanakkale | Female | 72 | 2020-03-18 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429864 | Sakarya | Male | 33 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429863 | Sakarya | Female | 42 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429862 | Ankara | Male | 65 | 2020-03-22 | 2020-04-24 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429861 | Ankara | Male | 48 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428723 | Aksaray | Male | 48 | 2020-03-22 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428722 | Balıkesir | Female | 37 | 2020-03-22 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428721 | Ankara | Male | NA | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428720 | Ankara | Female | 35 | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428719 | Siirt | Male | 52 | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428718 | Kocaeli | Male | 35 | 2020-03-19 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428717 | Kocaeli | Male | 38 | 2020-03-19 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428716 | Ankara | Female | 62 | 2020-03-18 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428715 | Nevşehir | Female | 55 | 2020-03-18 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428714 | Kastamonu | Male | 60 | 2020-03-18 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428713 | Ankara | Female | NA | 2020-03-18 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428712 | Karaman | Male | 72 | 2020-03-17 | 2020-04-21 | Patient travelled to France | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428368 | İstanbul | Female | 49 | 2020-04-16 | 2020-04-20 | NA | IlluminaNextSeq Assembly: BWA-MEM1,750x coverage |
| EPI_ISL_428346 | İstanbul | Male | 49 | 2020-04-17 | 2020-04-20 | NA | Illumina Next Seq assembly: BWA-MEM 2,350x coverage |
| EPI_ISL_427391 | İstanbul | Male | 51 | 2020-04-13 | 2020-04-18 | NA | Illumina Next Seq assembly: BWA-MEM 0.7.17.15,845x coverage |
| EPI_ISL_424366 | Kayseri | Male | 82 | 2020-03-17 | 2020-04-13 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 26,200x coverage |
| EPI_ISL_417413 | NA | Female | 27 | 2020-03-17 | 2020-03-25 | NA | Nanopore MinIONGeneious Prime245X coverage |
| EPI_ISL_435057 | Adıyaman | Male | 80 | 2020-04-09 | 2020-05-02 | NA | Oxford Nanopore MinION assembly: Geneious Prime 2020.1.240X coverage |
| EPI_ISL_437304 | Nevşehir | Female | 54 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437305 | Kocaeli | Female | 66 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437306 | Kocaeli | Male | 30 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437307 | Mardin | Female | 19 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437308 | Ankara | Male | 54 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437309 | Ankara | Female | 58 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437310 | Ankara | Male | NA | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437311 | Ankara | Male | 61 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437312 | Kocaeli | Male | 52 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437313 | Kocaeli | Male | 49 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437314 | Ankara | Male | 62 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437315 | Ankara | Female | 33 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437316 | Denizli | Female | NA | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437317 | Ankara | Female | 58 | 2020-03-27 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437318 | Ankara | Male | 31 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437319 | Kocaeli | Male | 47 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437320 | İstanbul | Female | 41 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437321 | İstanbul | Female | 25 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437322 | Ankara | Female | 62 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437323 | İstanbul | Male | 41 | 2020-03-19 | 2020-05-08 | Patient travelled toTaiwan | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437324 | İstanbul | Male | 27 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437325 | İstanbul | Male | 44 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437326 | İstanbul | Male | 38 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437327 | Ağrı | Female | 35 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437328 | Tekirdağ | Female | 35 | 2020-03-19 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437329 | Ankara | Male | 31 | 2020-03-19 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437330 | Tokat | Male | 20 | 2020-03-19 | 2020-05-08 | Health worker | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437331 | Ankara | Female | 59 | 2020-03-25 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437332 | İstanbul | Male | 50 | 2020-03-18 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437333 | Ankara | Male | 64 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437334 | Ankara | Female | 64 | 2020-03-24 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437335 | Denizli | Female | 79 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
Table .
Supplementary Table2. Nucleotide substitutions present in 62 SARS-CoV-2 viral genomes fromTurkey (submitted to GISAID between March 25th and May 22nd 2020) compared to the SARS-CoV-2 reference genome NC 045512.1. The nucleotide positions are given starting from 5’UTR. The star mark (*) indicates mutations that are only seen in the corresponding viral isolate.
| GISAID_sample accession ID | Nucleotide substitution at the given position | Corresponding viral gene | Corresponding viral gene product |
|---|---|---|---|
| EPI_ISL_417413 | *T580A | ORF1ab | Leader protein |
| *G779C | |||
| *T946A | ORF1ab | Nsp2 | |
| *T1100G | |||
| *C1101T | |||
| *A1106T | |||
| *A1119C | |||
| *A1134T | |||
| *G1156A | |||
| *G1210A | |||
| *C1225A | |||
| *T1359C | |||
| G1397A | |||
| *C1420T | |||
| *G1470A | |||
| *C1473T | |||
| *A1475C | |||
| *G2250A | |||
| C2455T | |||
| *A2475T | |||
| *G2549C | |||
| *T2586A | |||
| *G2591A | |||
| *G2612C | |||
| *G2715T | |||
| *A2932G | ORF1ab | Nsp3 | |
| C3117T | |||
| *G3146 C | |||
| C3787T | |||
| C4084T | |||
| *C7392T | |||
| *C11232T | ORF1ab | Nsp6 | |
| *G11234A | |||
| *C13476T | ORF1ab | RNA-dependent RNA polymerase | |
| *C13492T | |||
| *C14286T | |||
| *G14310A | |||
| *T14394A | |||
| *C14407A | |||
| *G14430A | |||
| *G14443T | |||
| *T14682G | |||
| *G14710A | |||
| *T14740C | |||
| *C14763A | |||
| *G14773T | |||
| *T14808A | |||
| *C15101A | |||
| *T15119A | |||
| *G15958A | |||
| *C19763A | ORF1ab | EndoRNAse | |
| *T26396A | E | Envelope | |
| *T26551C | M | Membrane glycoprotein | |
| *C26753T | |||
| C27103T | |||
| G28109T | ORF8 | ORF8 protein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_424366 | G1397A | ORF1ab | Nsp2 |
| G11083T | ORF1ab | Nsp6 | |
| *G23876A | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| *C29563T | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_427391 | C2113T | ORF1ab | Nsp2 |
| *C2997T | ORF1ab | Nsp3 | |
| C3037T | |||
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428368 | C3037T | ORF1ab | Nsp3 |
| G11083T | ORF1ab | Nsp6 | |
| *C12809T | ORF1ab | Nsp9 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_428346 | C2113T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’exonuclease | |
| *G21452T | ORF1ab | 2’-O-Ribose methyltransferase | |
| A23403G | S | Spike protein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428717 | C3037T | ORF1ab | Nsp3 |
| C12741T | ORF1ab | Nsp8 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| *C21304A | ORF1ab | 2’-O-Ribose methyltransferase | |
| *G21305A | |||
| A23403G | S | Spike protein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| *C28054T | ORF8 | ORF8 protein | |
| EPI_ISL_428716 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_428719 | C3037T | ORF1ab | Nsp3 |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428718 | C8782T | ORF1ab | Nsp4 |
| *G14122T | ORF1ab | RNA-dependent RNA polymerase | |
| G28878A | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428720 | C3037T | ORF1ab | Nsp3 |
| *G12248T | ORF1ab | Nsp8 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *T23559A | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_428722 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| C12741T | ORF1ab | Nsp9 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428721 | C3037T | ORF1ab | Nsp3 |
| C14178T | ORF1ab | RNA-dependent RNA polymerase | |
| C14408T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| G26718T | M | Membrane glycoprotein | |
| C26735T | |||
| EPI_ISL_428723 | G881A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_428713 | G1397A | ORF1ab | Nsp2 |
| C2455T | |||
| C3117T | ORF1ab | Nsp3 | |
| C3787T | |||
| C4084T | |||
| *C4524T | |||
| G11083T | ORF1ab | Nsp6 | |
| G28109T | ORF8 | ORF8 protein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428712 | C2416T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| G8371T | |||
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428715 | C3037T | ORF1ab | Nsp3 |
| C14178T | ORF1ab | RNA-dependent RNA polymerase | |
| C14408T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| G26718T | M | Membrane glycoprotein | |
| C26735T | |||
| EPI_ISL_428714 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429871 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429870 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C19170T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *C25275T | |||
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429873 | *C1437T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429872 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429862 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A20268G | ORF1ab | endoRNAse | |
| A23403G | S | Spike glycoprotein | |
| EPI_ISL_429861 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_429864 | *G944A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429863 | G881A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_429866 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429865 | G1397A | ORF1ab | Nsp2 |
| *C7834T | ORF1ab | Nsp3 | |
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| *C26340T | E | Envelope | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429868 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| *C11074T | ORF1ab | Nsp6 | |
| G11083T | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429867 | C884T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429869 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437304 | C241T | 5’ UTR | |
| C2113T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *C25549T | ORF3a | ORF3a protein | |
| G25563T | |||
| EPI_ISL_437305 | C241T | 5’UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437306 | G1397A | ORF1ab | Nsp2 |
| G8653T | ORF1ab | Nsp4 | |
| *C8683A | |||
| G11083T | ORF1ab | Nsp6 | |
| A23403G | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| ORF1ab | Nsp2 | ||
| EPI_ISL_437307 | *C1314T | ORF1ab | Nsp2 |
| G1397A | |||
| *T6202A | ORF1ab | Nsp2 | |
| *C8964T | ORF1ab | Nsp4 | |
| *C10202T | ORF1ab | 3C-like proteinase | |
| 1G1083T | ORF1ab | Nsp6 | |
| C13481T | ORF1ab | Nsp10 | |
| *C16247T | ORF1ab | Helicase | |
| *C24865T | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437308 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C15240T | |||
| T19839C | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C26256T | E | Envelope | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437309 | C241T | 5’ UTR | |
| C1059T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| C3903T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C16616T | ORF1ab | Helicase | |
| A23403G | S | Spike glycoprotein | |
| *A23734T | |||
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437310 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437311 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437312 | C1397T | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437313 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437314 | G1397A | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| C5736T | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| C23874T | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437315 | C1059T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437316 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C5736 T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A20268G | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C23874T | |||
| EPI_ISL_437317 | C24T | 5’ UTR | |
| C8782T | ORF1ab | Nsp4 | |
| *G22468T | S | Spike glycoprotein | |
| *G25314T | |||
| *T28144C | ORF8 | ORF8 protein | |
| G28878A | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437318 | G1397A | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| *C5477T | |||
| C5736T | |||
| *C6402T | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437319 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| *G19285A | ORF1ab | 3’ to 5’ exonuclease | |
| C21789T | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437320 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437321 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437322 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437323 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437324 | G1397A | ORF1ab | Nsp2 |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437325 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| C5736T | ORF1ab | Nsp3 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C21789T | S | Spike glycoprotein | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437326 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C5736T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26256T | E | Envelope | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437327 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437328 | C241T | 5’ UTR | |
| *C1825T | ORF1ab | Nsp2 | |
| C2113T | |||
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437329 | C228T | 5’ UTR | |
| C5736T | ORF1ab | Nsp3 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| C13481T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| G26720C | M | Membrane glycoprotein | |
| EPI_ISL_437330 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| *C5826A | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA dependent RNA polymerase | |
| C18877T | ORF1ab | 3’to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26549T | M | Membrane glycoprotein | |
| EPI_ISL_437331 | C228T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| *C12700T | ORF1ab | Nsp9 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C19484T | ORF1ab | 3’to 5’ exonuclease | |
| A20268G | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C23874T | |||
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| C29741T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437332 | C228T | 5’ UTR | |
| C2416T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| G8371T | ORF1ab | Nsp3 | |
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| G25563T | ORF3a | ORF3a protein | |
| C26549T | M | Membrane glycoprotein | |
| C27103T | |||
| EPI_ISL_437333 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *T15102C | |||
| T19839C | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437334 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| C3903T | ORF1ab | Nsp3 | |
| A9514G | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| *A13376G | ORF1ab | Nsp10 | |
| C13481T | ORF1ab | RNA-dependent RNA polymerase | |
| C19484T | ORF1ab | 3’to 5’ exonuclease | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437335 | C228T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C3903T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| *A27354G | ORF6 | ORF6 protein |
Footnotes
Worldometers (2020). Global COVID-19 statistics [online]. Website https://www.worldometers.info/coronavirus/ [accessed 05 May 2020].
T.C. Sağlık Bakanlığı (2020). Türkiye’deki Güncel Durum [online]. Website https://covid19.saglik.gov.tr/ [accessed 05 May 2020].
GISAID (2020). Website https://www.gisaid.org/ [accessed 24 April 2020].
NCBI (2020). GeneBank Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome [online]. Website https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2 [accessed 24 April 2020].
MAFFT (2020). Multiple alignment program for amino acid or nucleotide sequences [online]. Website https://mafft.cbrc.jp/alignment/software/ [accessed 25 April 2020].
Issues with SARS-CoV-2 sequencing data (2020). [online]. Website https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473. [Accessed 5 May 2020]
References
- Evolution and molecular characteristics of SARS-CoV-2 genome. BioRxiv (preprint. doi: 10. 2020. [DOI] [PMC free article] [PubMed]
- Benvenuto D Angeletti S Giovanetti M Bianchi M Pascarella S Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. Journal of Infection. 2020;03 doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. doi: 10. 2020. [DOI] [PMC free article] [PubMed]
- Eden JS Rockett R Carter I Rahman H De Ligt An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. Virus Evolution. 2020;6:1. doi: 10.1093/ve/veaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S Buckland-Merrett G Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges. 2017;1:33. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RdRp mutations areassociated with SARS-CoV-2 genome evolution. BioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- Forster P Forster L Renfrew C Forster M Phylogenetic network analysis of SARS- CoV-2 genomes. Proceedings of the National Academy of Sciences. 2020;117:9241. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigidity of the outer shell. 2020. [DOI] [PMC free article] [PubMed]
- predicted by a protein intrinsic disorder model sheds light on the COVID-19 (Wuhan-2019-nCoV) infectivity. Biomolecules. 10:2019. doi: 10.3390/biom10020331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE Baker SC Baric RS De Groot RJ Drosten C The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Structure of replicating SARS-CoV-2 polymerase. BioRxiv (preprint. doi: 10.1038/s41586-020-2368-8. 2020. [DOI] [PubMed]
- Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. doi: 10. 2020.
- Li J Li Z Cui X Wu C Bayesian phylodynamic inference on the temporal evolution and global transmission of SARS-CoV-2. doi: 10. 2020;04:1. doi: 10.1016/j.jinf.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J Plessis L Hill Z Kang V M Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181:997. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X Liu Y Lei X Li P Mi D Characterization of spike glycoprotein of SARS- CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications. 2020;11:1620–1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M Marini B Benedetti F Giudici F Mauro E Emerging SARS-CoV 2 mutation hot spots include a novel RNA - dependent - RNA polymerase variant. Journal of Translational Medicine. 2020;18:1. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS Mou H Smits SL Dekkers DHW Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman SU Shafique L Ihsan A Liu Q Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. 2020;10:1. doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y McCauley J GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22:2. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C Liu Z Chen Z Huang X Xu M The establishment of reference sequence for SARS-CoV-2 and variation analysis. Journal of Medical Virology . 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
General information taken from GISAID about the SARS-CoV-2 sequences used in this study. (NA: Not available)
| GISAID ID | Location | Sex | Age | Collection date | Submission date | Travel history/other | Sequencing technology |
|---|---|---|---|---|---|---|---|
| EPI_ISL_429873 | Kocaeli | Male | 71 | 2020-03-23 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429872 | Kocaeli | Female | 50 | 2020-03-25 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429871 | Ankara | Male | 77 | 2020-03-23 | 2020-04-24 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429870 | Sakarya | Female | 57 | 2020-03-22 | 2020-04024 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429869 | Konya | Female | 59 | 2020-03-17 | 2020-04-24 | Patient travelled to Saudi Arabia | IlluminaMiseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429868 | Eskişehir | Female | 79 | 2020-03-17 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429867 | Balıkesir | Female | 72 | 2020-03-17 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429866 | Afyon | Female | 52 | 2020-03-16 | 2020-04-24 | Patient travelled to Saudi Arabia | IlluminaMiseq Assembly: Burrows-Wheeler Aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429865 | Çanakkale | Female | 72 | 2020-03-18 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429864 | Sakarya | Male | 33 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429863 | Sakarya | Female | 42 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429862 | Ankara | Male | 65 | 2020-03-22 | 2020-04-24 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_429861 | Ankara | Male | 48 | 2020-03-22 | 2020-04-24 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428723 | Aksaray | Male | 48 | 2020-03-22 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428722 | Balıkesir | Female | 37 | 2020-03-22 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428721 | Ankara | Male | NA | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428720 | Ankara | Female | 35 | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428719 | Siirt | Male | 52 | 2020-03-21 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428718 | Kocaeli | Male | 35 | 2020-03-19 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428717 | Kocaeli | Male | 38 | 2020-03-19 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428716 | Ankara | Female | 62 | 2020-03-18 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428715 | Nevşehir | Female | 55 | 2020-03-18 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428714 | Kastamonu | Male | 60 | 2020-03-18 | 2020-04-21 | Patient travelled to Saudi Arabia | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428713 | Ankara | Female | NA | 2020-03-18 | 2020-04-21 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428712 | Karaman | Male | 72 | 2020-03-17 | 2020-04-21 | Patient travelled to France | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 1,000x coverage |
| EPI_ISL_428368 | İstanbul | Female | 49 | 2020-04-16 | 2020-04-20 | NA | IlluminaNextSeq Assembly: BWA-MEM1,750x coverage |
| EPI_ISL_428346 | İstanbul | Male | 49 | 2020-04-17 | 2020-04-20 | NA | Illumina Next Seq assembly: BWA-MEM 2,350x coverage |
| EPI_ISL_427391 | İstanbul | Male | 51 | 2020-04-13 | 2020-04-18 | NA | Illumina Next Seq assembly: BWA-MEM 0.7.17.15,845x coverage |
| EPI_ISL_424366 | Kayseri | Male | 82 | 2020-03-17 | 2020-04-13 | NA | Illumina Miseq assembly: Burrows-Wheeler aligner v.07.17-r1188 26,200x coverage |
| EPI_ISL_417413 | NA | Female | 27 | 2020-03-17 | 2020-03-25 | NA | Nanopore MinIONGeneious Prime245X coverage |
| EPI_ISL_435057 | Adıyaman | Male | 80 | 2020-04-09 | 2020-05-02 | NA | Oxford Nanopore MinION assembly: Geneious Prime 2020.1.240X coverage |
| EPI_ISL_437304 | Nevşehir | Female | 54 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437305 | Kocaeli | Female | 66 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437306 | Kocaeli | Male | 30 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437307 | Mardin | Female | 19 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437308 | Ankara | Male | 54 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437309 | Ankara | Female | 58 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437310 | Ankara | Male | NA | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437311 | Ankara | Male | 61 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437312 | Kocaeli | Male | 52 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437313 | Kocaeli | Male | 49 | 2020-03-27 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437314 | Ankara | Male | 62 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437315 | Ankara | Female | 33 | 2020-03-26 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437316 | Denizli | Female | NA | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437317 | Ankara | Female | 58 | 2020-03-27 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437318 | Ankara | Male | 31 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437319 | Kocaeli | Male | 47 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437320 | İstanbul | Female | 41 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437321 | İstanbul | Female | 25 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437322 | Ankara | Female | 62 | 2020-03-19 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437323 | İstanbul | Male | 41 | 2020-03-19 | 2020-05-08 | Patient travelled toTaiwan | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437324 | İstanbul | Male | 27 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437325 | İstanbul | Male | 44 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437326 | İstanbul | Male | 38 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437327 | Ağrı | Female | 35 | 2020-03-19 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437328 | Tekirdağ | Female | 35 | 2020-03-19 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437329 | Ankara | Male | 31 | 2020-03-19 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437330 | Tokat | Male | 20 | 2020-03-19 | 2020-05-08 | Health worker | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437331 | Ankara | Female | 59 | 2020-03-25 | 2020-05-08 | Patient travelled to Saudi Arabia | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437332 | İstanbul | Male | 50 | 2020-03-18 | 2020-05-08 | Patient travelled to Iran | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437333 | Ankara | Male | 64 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437334 | Ankara | Female | 64 | 2020-03-24 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
| EPI_ISL_437335 | Denizli | Female | 79 | 2020-03-25 | 2020-05-08 | NA | Oxford Nanopore GridION assembly: Geneious Prime1000X coverage |
Table .
Supplementary Table2. Nucleotide substitutions present in 62 SARS-CoV-2 viral genomes fromTurkey (submitted to GISAID between March 25th and May 22nd 2020) compared to the SARS-CoV-2 reference genome NC 045512.1. The nucleotide positions are given starting from 5’UTR. The star mark (*) indicates mutations that are only seen in the corresponding viral isolate.
| GISAID_sample accession ID | Nucleotide substitution at the given position | Corresponding viral gene | Corresponding viral gene product |
|---|---|---|---|
| EPI_ISL_417413 | *T580A | ORF1ab | Leader protein |
| *G779C | |||
| *T946A | ORF1ab | Nsp2 | |
| *T1100G | |||
| *C1101T | |||
| *A1106T | |||
| *A1119C | |||
| *A1134T | |||
| *G1156A | |||
| *G1210A | |||
| *C1225A | |||
| *T1359C | |||
| G1397A | |||
| *C1420T | |||
| *G1470A | |||
| *C1473T | |||
| *A1475C | |||
| *G2250A | |||
| C2455T | |||
| *A2475T | |||
| *G2549C | |||
| *T2586A | |||
| *G2591A | |||
| *G2612C | |||
| *G2715T | |||
| *A2932G | ORF1ab | Nsp3 | |
| C3117T | |||
| *G3146 C | |||
| C3787T | |||
| C4084T | |||
| *C7392T | |||
| *C11232T | ORF1ab | Nsp6 | |
| *G11234A | |||
| *C13476T | ORF1ab | RNA-dependent RNA polymerase | |
| *C13492T | |||
| *C14286T | |||
| *G14310A | |||
| *T14394A | |||
| *C14407A | |||
| *G14430A | |||
| *G14443T | |||
| *T14682G | |||
| *G14710A | |||
| *T14740C | |||
| *C14763A | |||
| *G14773T | |||
| *T14808A | |||
| *C15101A | |||
| *T15119A | |||
| *G15958A | |||
| *C19763A | ORF1ab | EndoRNAse | |
| *T26396A | E | Envelope | |
| *T26551C | M | Membrane glycoprotein | |
| *C26753T | |||
| C27103T | |||
| G28109T | ORF8 | ORF8 protein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_424366 | G1397A | ORF1ab | Nsp2 |
| G11083T | ORF1ab | Nsp6 | |
| *G23876A | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| *C29563T | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_427391 | C2113T | ORF1ab | Nsp2 |
| *C2997T | ORF1ab | Nsp3 | |
| C3037T | |||
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428368 | C3037T | ORF1ab | Nsp3 |
| G11083T | ORF1ab | Nsp6 | |
| *C12809T | ORF1ab | Nsp9 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_428346 | C2113T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’exonuclease | |
| *G21452T | ORF1ab | 2’-O-Ribose methyltransferase | |
| A23403G | S | Spike protein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428717 | C3037T | ORF1ab | Nsp3 |
| C12741T | ORF1ab | Nsp8 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| *C21304A | ORF1ab | 2’-O-Ribose methyltransferase | |
| *G21305A | |||
| A23403G | S | Spike protein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| *C28054T | ORF8 | ORF8 protein | |
| EPI_ISL_428716 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_428719 | C3037T | ORF1ab | Nsp3 |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428718 | C8782T | ORF1ab | Nsp4 |
| *G14122T | ORF1ab | RNA-dependent RNA polymerase | |
| G28878A | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428720 | C3037T | ORF1ab | Nsp3 |
| *G12248T | ORF1ab | Nsp8 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *T23559A | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_428722 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| C12741T | ORF1ab | Nsp9 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428721 | C3037T | ORF1ab | Nsp3 |
| C14178T | ORF1ab | RNA-dependent RNA polymerase | |
| C14408T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| G26718T | M | Membrane glycoprotein | |
| C26735T | |||
| EPI_ISL_428723 | G881A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_428713 | G1397A | ORF1ab | Nsp2 |
| C2455T | |||
| C3117T | ORF1ab | Nsp3 | |
| C3787T | |||
| C4084T | |||
| *C4524T | |||
| G11083T | ORF1ab | Nsp6 | |
| G28109T | ORF8 | ORF8 protein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_428712 | C2416T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| G8371T | |||
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_428715 | C3037T | ORF1ab | Nsp3 |
| C14178T | ORF1ab | RNA-dependent RNA polymerase | |
| C14408T | |||
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| G26718T | M | Membrane glycoprotein | |
| C26735T | |||
| EPI_ISL_428714 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429871 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429870 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C19170T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *C25275T | |||
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429873 | *C1437T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429872 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429862 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A20268G | ORF1ab | endoRNAse | |
| A23403G | S | Spike glycoprotein | |
| EPI_ISL_429861 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_429864 | *G944A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_429863 | G881A | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_429866 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429865 | G1397A | ORF1ab | Nsp2 |
| *C7834T | ORF1ab | Nsp3 | |
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| *C26340T | E | Envelope | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429868 | C884T | ORF1ab | Nsp2 |
| G1397A | |||
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| *C11074T | ORF1ab | Nsp6 | |
| G11083T | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_429867 | C884T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| EPI_ISL_429869 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C26735T | M | Membrane glycoprotein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437304 | C241T | 5’ UTR | |
| C2113T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| *C25549T | ORF3a | ORF3a protein | |
| G25563T | |||
| EPI_ISL_437305 | C241T | 5’UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437306 | G1397A | ORF1ab | Nsp2 |
| G8653T | ORF1ab | Nsp4 | |
| *C8683A | |||
| G11083T | ORF1ab | Nsp6 | |
| A23403G | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| ORF1ab | Nsp2 | ||
| EPI_ISL_437307 | *C1314T | ORF1ab | Nsp2 |
| G1397A | |||
| *T6202A | ORF1ab | Nsp2 | |
| *C8964T | ORF1ab | Nsp4 | |
| *C10202T | ORF1ab | 3C-like proteinase | |
| 1G1083T | ORF1ab | Nsp6 | |
| C13481T | ORF1ab | Nsp10 | |
| *C16247T | ORF1ab | Helicase | |
| *C24865T | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437308 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C15240T | |||
| T19839C | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C26256T | E | Envelope | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437309 | C241T | 5’ UTR | |
| C1059T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| C3903T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *C16616T | ORF1ab | Helicase | |
| A23403G | S | Spike glycoprotein | |
| *A23734T | |||
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437310 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| A23403G | |||
| G25563T | ORF3a | ORF3a protein | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437311 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437312 | C1397T | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437313 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437314 | G1397A | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| C5736T | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| C23874T | S | Spike glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437315 | C1059T | ORF1ab | Nsp2 |
| C3037T | ORF1ab | Nsp3 | |
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437316 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C5736 T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A20268G | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C23874T | |||
| EPI_ISL_437317 | C24T | 5’ UTR | |
| C8782T | ORF1ab | Nsp4 | |
| *G22468T | S | Spike glycoprotein | |
| *G25314T | |||
| *T28144C | ORF8 | ORF8 protein | |
| G28878A | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437318 | G1397A | ORF1ab | Nsp2 |
| T5182C | ORF1ab | Nsp3 | |
| *C5477T | |||
| C5736T | |||
| *C6402T | |||
| G8653T | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437319 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| *G19285A | ORF1ab | 3’ to 5’ exonuclease | |
| C21789T | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437320 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437321 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C26549T | M | Membrane glycoprotein | |
| G26720C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437322 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437323 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437324 | G1397A | ORF1ab | Nsp2 |
| G8653T | ORF1ab | Nsp4 | |
| C10702T | ORF1ab | 3C-like proteinase | |
| G11083T | ORF1ab | Nsp6 | |
| A22964G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| T28688C | N | Nucleocapsid phosphoprotein | |
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437325 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| C5736T | ORF1ab | Nsp3 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| C21789T | S | Spike glycoprotein | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437326 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C5736T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| C22444T | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26256T | E | Envelope | |
| C28854T | N | Nucleocapsid phosphoprotein | |
| EPI_ISL_437327 | C241T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| G11083T | ORF1ab | Nsp6 | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437328 | C241T | 5’ UTR | |
| *C1825T | ORF1ab | Nsp2 | |
| C2113T | |||
| C3037T | ORF1ab | Nsp3 | |
| C7765T | |||
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C17690T | ORF1ab | Helicase | |
| C18877T | ORF1ab | 3’to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| EPI_ISL_437329 | C228T | 5’ UTR | |
| C5736T | ORF1ab | Nsp3 | |
| G9479T | ORF1ab | Nsp4 | |
| A9514G | |||
| C13481T | ORF1ab | RNA-dependent RNA polymerase | |
| C18877T | ORF1ab | 3’ to 5’ exonuclease | |
| G26720C | M | Membrane glycoprotein | |
| EPI_ISL_437330 | C241T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| *C5826A | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA dependent RNA polymerase | |
| C18877T | ORF1ab | 3’to 5’ exonuclease | |
| A23403G | S | Spike glycoprotein | |
| G25563T | ORF3a | ORF3a protein | |
| C26549T | M | Membrane glycoprotein | |
| EPI_ISL_437331 | C228T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| *C12700T | ORF1ab | Nsp9 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| C19484T | ORF1ab | 3’to 5’ exonuclease | |
| A20268G | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C23874T | |||
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| C29741T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437332 | C228T | 5’ UTR | |
| C2416T | ORF1ab | Nsp2 | |
| C3037T | ORF1ab | Nsp3 | |
| G8371T | ORF1ab | Nsp3 | |
| G11083T | ORF1ab | Nsp6 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| G25563T | ORF3a | ORF3a protein | |
| C26549T | M | Membrane glycoprotein | |
| C27103T | |||
| EPI_ISL_437333 | C3037T | ORF1ab | Nsp3 |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| *T15102C | |||
| T19839C | ORF1ab | EndoRNase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| G28881A | N | Nucleocapsid phosphoprotein | |
| G28882A | |||
| G28883C | |||
| EPI_ISL_437334 | C228T | 5’ UTR | |
| G1397A | ORF1ab | Nsp2 | |
| C3903T | ORF1ab | Nsp3 | |
| A9514G | ORF1ab | Nsp4 | |
| G11083T | ORF1ab | Nsp6 | |
| *A13376G | ORF1ab | Nsp10 | |
| C13481T | ORF1ab | RNA-dependent RNA polymerase | |
| C19484T | ORF1ab | 3’to 5’ exonuclease | |
| G26720C | M | Membrane glycoprotein | |
| T28688C | N | Nucleocapsid phosphoprotein | |
| T28835C | |||
| G29742T | 3’UTR stem loop II like motif | ||
| EPI_ISL_437335 | C228T | 5’ UTR | |
| C3037T | ORF1ab | Nsp3 | |
| C3903T | ORF1ab | Nsp3 | |
| C14408T | ORF1ab | RNA-dependent RNA polymerase | |
| A23403G | S | Spike glycoprotein | |
| C26549T | M | Membrane glycoprotein | |
| T26551C | |||
| *A27354G | ORF6 | ORF6 protein |