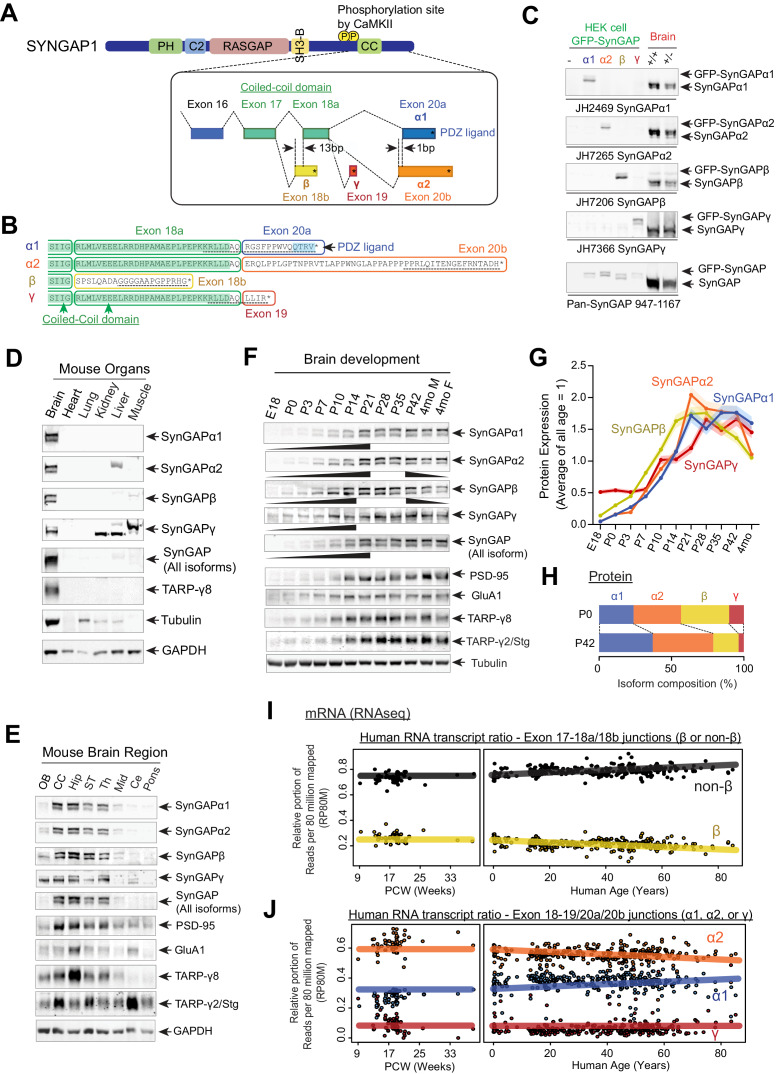
Figure 1. SynGAP isoforms are differentially expressed during brain development.
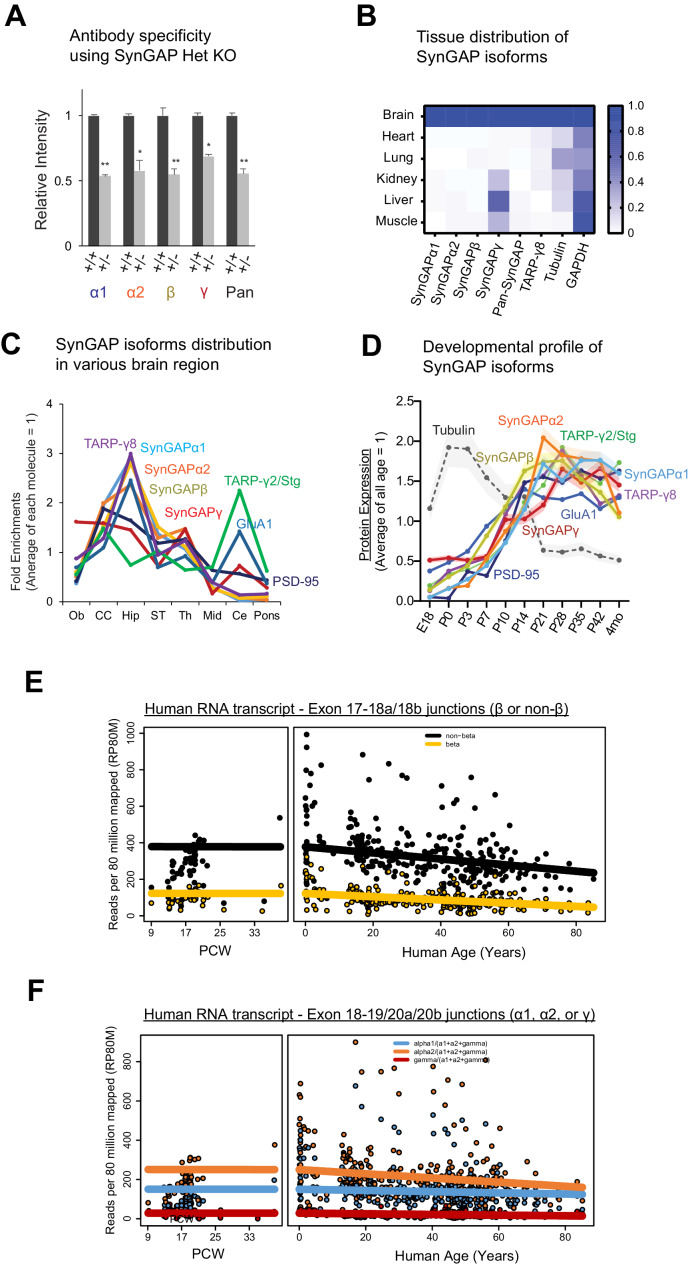
(A) Schematic of SYNGAP1 splicing at the C-terminus. SYNGAP1 is alternatively spliced within exons 18–20 to generate four unique C-terminal isoforms designated as α1, α2, β, and γ. (B) C-terminal amino-acid sequences of SynGAP isoforms encoding select protein domains. Coil-Coil domain (yellow) and PDZ ligand-binding domain (blue). Targeted epitopes of isoform-specific SynGAP antibodies (JH2469, JH7265, JH7206, and JH7366) are indicated as dotted lines. (C) Specificity of SynGAP isoform-specific antibodies. Immunoblots of SynGAP isoform expression in lysates prepared from HEK 293 T cells expressing individual GFP-tagged SynGAP isoforms and lysates prepared from brain tissue obtained from WT and Syngap1 +/- mice were shown. Quantification of relative SynGAP isoform levels with respect to total SynGAP expression measured from immunoblot were shown in Figure 1—figure supplement 1A. Two-way ANOVA followed by Tukey's post hoc test (Isoform F(4,30) = 1.900; p=0.13, Genotype F(1,30) = 451.2; p<0.001, Interaction F(4,30)=1.900; p=0.13, n = 4 each condition) was performed. Error bar indicates ± SEM. (D) Endogenous expression and distribution of SynGAP isoforms in various organs. Immunoblots of qualitative distribution of SynGAP isoforms in lysates prepared from various organ tissues of WT mice were shown. Asterisks indicate non-specific bands that are also detected in tissue from knockout mice. Two-way ANOVA followed by Tukey's post hoc test (Tissue F(5,144) = 1433; p<0.0001, Isoform F(7,144) = 229.3; p<0.0001, Interaction F(35,144) = 25.45; p<0.0001, n = 4 each condition) was performed. Heat map of immunoblots was displayed in Figure 1—figure supplement 1B. The amount of protein in the brain is standardized as 1.0. (E) Western blot of endogenous levels of individual SynGAP isoforms and other synaptic proteins in lysates prepared from several brain regions obtained from WT and Syngap1 +/- mice. (OB: Olfactory bulb, CC: Cerebral cortex, Hip: Hippocampus, ST: Striatum, Th: Thalamus, Mid: Midbrain, Ce: Cerebellum). Two-way ANOVA followed by Tukey's post hoc test (Brain regions F(7, 264)=1048; p<0.0001, Molecules F(10,264) = 8.0 x 10 −12; p>0.9999, Interaction F(70.264) = 59.06; p<0.0001, n = 4 each condition) was performed. Graph showing the mean values of each signal was displayed in Figure 1—figure supplement 1C. (F–H) Developmental expression profiles of individual SynGAP isoforms and related synaptic proteins. (F) Immunoblots of SynGAP isoform expression measured in forebrain tissue lysates prepared from WT and Syngap1 +/- mice at different developmental ages. (G) Quantification of immunoblots representing relative enrichments along developmental stage. The mean values of each signal were plotted in the graph. (H) Quantification of absolute SynGAP isoform abundance at P0 and P42 from C and G. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Developmental stage F(10,330) = 397.4; p<0.0001, Molecule F(9,330) = 2.116; p=0.027, Interaction F(90,330) = 26.18; p<0.0001, n = 4 each condition) was performed. (I) mRNA expression of the β and non-β SYNGAP1 isoforms across age in human dorsolateral prefrontal cortex. The relative portion of RNAseq reads spanning the exon 17–18 junction supporting either isoform was plotted against human age (post-conception weeks and years) with a linear regression. (J) mRNA expression of the α1, α2, and γ SYNGAP1 isoforms across age. The relative portion of RNAseq reads spanning the exon 18–19 junction (γ) or 18–20 (α1, α2) junction supporting each isoform was plotted against human age. Reads per 80 million mapped (RP80M) of RNAseq data are shown in Figure 1—figure supplement 1E,F.