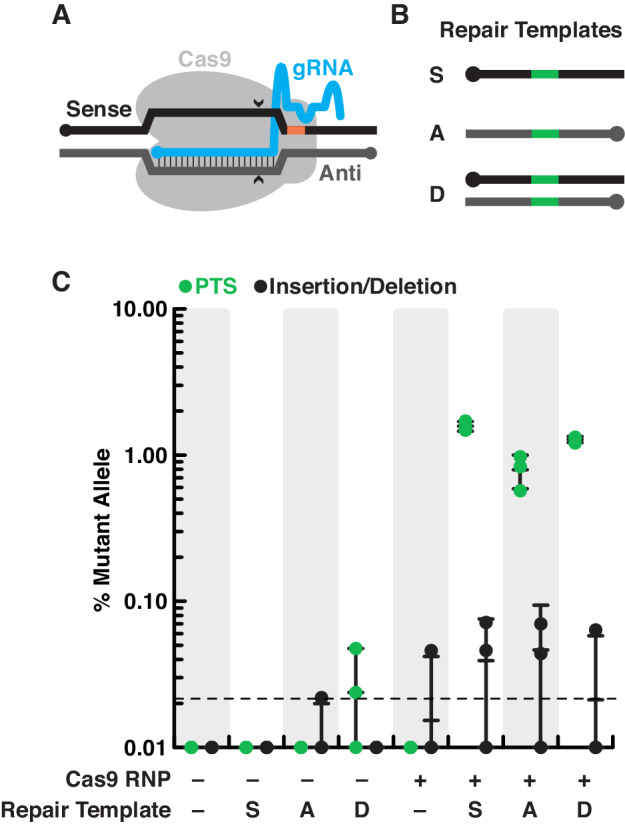
Figure 4. S. rosetta preferentially introduces genome-edited mutations from DNA templates.
(A) Schematic of a gRNA targeting SpCas9 to a genomic locus of interest. A gRNA (cyan, knobs indicate 5’ ends) that encodes a 20 nt targeting sequence from the sense strand of a genomic locus (black) hybridizes with the antisense strand (dark gray). SpCas9 (light gray) introduces a double-stranded break at the genomic locus (carets), 3 bp upstream of a protospacer adjacent motif (PAM, orange). (B) We designed a panel of repair oligonucleotides to test the preferred substrates for repairing double-stranded breaks introduced by SpCas9 at rosetteles exon 4. Oligonucleotide repair templates containing the PTS sequence (green) were delivered as single-stranded DNA in the sense (S) or anti-sense (A) orientations and as a double-stranded template (D) to test which most efficiently templated DNA repair at the SpCas9 cleavage site. (C) SpCas9 stimulated the introduction of PTS mutations from DNA templates. Repair templates with a PTS (from panel B) were delivered in the presence and absence of SpCas9 (+/–). A ~ 450 bp fragment surrounding the rtlsPTS1 cleavage site was amplified from cells that had been transfected the previous day to prepare deep sequencing libraries for quantifying the frequency of PTS insertions (green) or insertions/deletions from error prone editing (black). Each experiment was performed three independent times (dots; mean and standard deviations are shown with lines). The dotted line indicates the limit of detection of the sequencing, based on a 6-base, randomized barcode. Upon transfection with the SpCas9 RNP, 10x more mutations from repair templates (1–2%, green dots) were detected than untemplated insertions or deletions (black dots). In the absence of SpCas9, mutations generated from a double-stranded template, but not single-stranded templates, were rarely (<0.1%) and unreliably (2 of 3 trials) found.