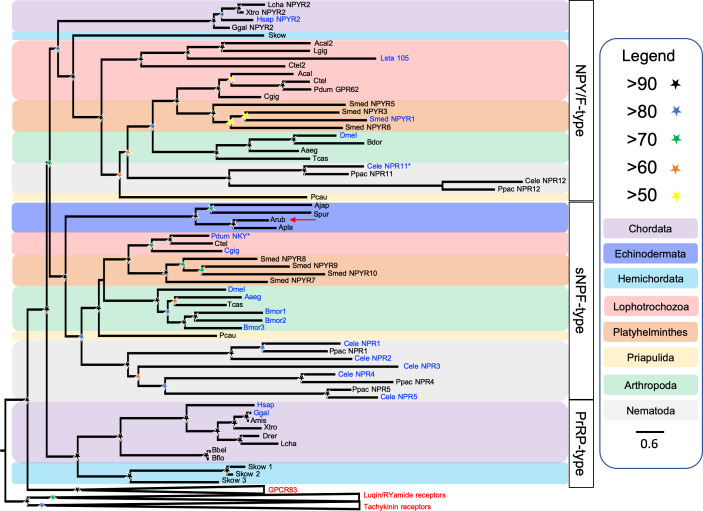
Figure 4. Phylogenetic tree showing that a candidate receptor for the A. rubens neuropeptide ArPrRP is an ortholog of protostome sNPF-type receptors.
The tree includes NPY/NPF-type receptors, chordate PrRP-type receptors and protostome sNPF-type receptors, with GPR83-type, luqin-type, and tachykinin-type receptors as outgroups to root the tree. Interestingly, the candidate receptor for the A. rubens neuropeptide ArPrRP (red arrow) and orthologs from other echinoderms are positioned in a clade comprising protostome sNPF-type receptors, whereas candidate receptors for PrRP-type peptides in the hemichordate S. kowalevskii are positioned in a clade containing chordate PrRP-type receptors. Note that NPY/NPF-type receptors form a distinct clade that includes an NPY/NPF-type receptor from the hemichordate S. kowalevskii, but no echinoderm receptors are present in this clade. The tree was generated in W-IQ-tree 1.0 using the Maximum likelihood method. The stars represent bootstrap support (1000 replicates, see legend) and the coloured backgrounds represent different taxonomic groups, as shown in the key. The names with text in blue represent the receptors for which ligands have been experimentally confirmed. The asterisks highlight receptors where the reported ligand is atypical when compared with ligands for receptors in the same clade. Species names are as follows: Aaeg (Aedes aegypti), Acal (Aplysia californica), Ajap (Apostichopus japonicus), Amis (Alligator mississippiensis), Apla (Acanthaster planci), Arub (Asterias rubens), Bbel (Branchiostoma belcheri), Bdor (Bactrocera dorsalis), Bflo (Branchiostoma floridae), Bmor (Bombyx mori), Cele (Caenorhabditis elegans), Cgig (Crassostrea gigas), Ctel (Capitella teleta), Dmel (Drosophila melanogaster), Drer (Danio rerio), Ggal (Gallus gallus), Hsap (Homo sapiens), Lcha (Latimeria chalumnae), Lgig (Lottia gigantea), Lsta (Lymnaea stagnalis), Pcau (Priapulus caudatus), Pdum (Platynereis dumerilii), Ppac (Pristionchus pacificus), Skow (Saccoglossus kowalevskii), Smed (Schmidtea mediterranea), Spur (Strongylocentrotus purpuratus), Tcas (Tribolium castaneum), Xtro (Xenopus tropicalis). The accession numbers of the sequences used for this phylogenetic tree are listed in Figure 4—source data 1.