Abstract
Background
Pathogen-based diagnostics for acute respiratory infection (ARI) have limited ability to detect etiology of illness. We previously showed that peripheral blood-based host gene expression classifiers accurately identify bacterial and viral ARI in cohorts of European and African descent. We determined classifier performance in a South Asian cohort.
Methods
Patients ≥15 years with fever and respiratory symptoms were enrolled in Sri Lanka. Comprehensive pathogen-based testing was performed. Peripheral blood ribonucleic acid was sequenced and previously developed signatures were applied: a pan-viral classifier (viral vs nonviral) and an ARI classifier (bacterial vs viral vs noninfectious).
Results
Ribonucleic acid sequencing was performed in 79 subjects: 58 viral infections (36 influenza, 22 dengue) and 21 bacterial infections (10 leptospirosis, 11 scrub typhus). The pan-viral classifier had an overall classification accuracy of 95%. The ARI classifier had an overall classification accuracy of 94%, with sensitivity and specificity of 91% and 95%, respectively, for bacterial infection. The sensitivity and specificity of C-reactive protein (>10 mg/L) and procalcitonin (>0.25 ng/mL) for bacterial infection were 100% and 34%, and 100% and 41%, respectively.
Conclusions
Previously derived gene expression classifiers had high predictive accuracy at distinguishing viral and bacterial infection in South Asian patients with ARI caused by typical and atypical pathogens.
Keywords: antimicrobial stewardship, diagnostic test, gene expression, respiratory tract infection, Sri Lanka
Host biomarkers can identify bacterial versus viral acute respiratory infection (ARI). Previously, we derived host gene expression classifiers that accurately identify bacterial versus viral ARI. Here, we show that our classifiers had high predictive accuracy in a South Asian cohort.
Acute respiratory infection (ARI) is a leading cause of illness globally [1, 2]. A large proportion of ARI is caused by viruses such as human rhinovirus/human enterovirus, influenza, and respiratory syncytial virus (RSV), but viral ARI remains one of the most common reasons for inappropriate antibiotic use worldwide [3–7]. The overuse of antibiotics in human healthcare is associated with an alarming rise in antimicrobial resistance [8]. If left uncurbed, antimicrobial-resistant infections are estimated to result in up to 10 million deaths annually by 2050 [9].
Challenges associated with ARI diagnosis are an important contributor to inappropriate antibiotic use [10]. Even with comprehensive testing, pathogen-based diagnostics such as culture and nucleic acid amplification may fail to detect an organism [11, 12]. Furthermore, the identification of an organism may represent asymptomatic shedding rather than infection [13]. In tropical and subtropical settings, diagnosis is further confounded by endemic febrile infections such as dengue, leptospirosis, and Q fever, which frequently present with respiratory symptoms [14–16]. Pathogen-based diagnosis of such illnesses generally requires laboratory infrastructure and skilled labor that are only available in reference laboratories, leading clinicians to err on the side of overtreating with antibiotics [17]. An ideal ARI diagnostic in tropical settings would thus cover the spectrum of endemic pathogens, while offering a timely result to inform decisions about antimicrobial use [17].
Given the limitations associated with pathogen-based diagnostics, host-based biomarkers that detect response to infection are an attractive complementary strategy for the diagnosis of ARI [18]. C-reactive protein (CRP) has been used for decades to diagnose the presence of bacterial infection, but it has been characterized by low specificity [19, 20]. Procalcitonin (PCT) has emerged as a biomarker specific for bacterial respiratory infection, but sensitivity and specificity have been found to be as low as 55% in some studies [21–23]. More importantly, PCT has not been widely evaluated in tropical and subtropical settings, and limited existing data suggest that PCT may not perform well at identifying bacterial infections such as leptospirosis and rickettsial infection [19].
Our team has developed peripheral blood-based gene expression classifiers that assess a patient’s response to respiratory infection across the transcriptome [24–26]. A pan-viral gene classifier consisting of 28 unique genes was initially developed to identify symptomatic viral ARI from other states. This classifier identified viral ARI from uninfected individuals with 97% accuracy and viral ARI from bacterial ARI with 80% accuracy [25]. A more complex model, known as the ARI classifier, was then developed to discriminate both bacterial and viral ARI states. The ARI classifier consists of 3 independent signatures for bacterial, viral, and noninfectious illnesses. All 3 are applied to a symptomatic individual such that the signature with the highest probability determines class assignment. This classifier discriminated bacterial, viral, and noninfectious states with an overall accuracy of 87% [26]. Both the pan-viral and ARI classifiers were derived using cohorts that included subjects of mostly white or black race in the United States and the United Kingdom, and who had infections caused by typical respiratory pathogens such as influenza, RSV, and Streptococcus pneumoniae. These classifiers have not been assessed in populations of diverse race and ethnicity and from geographic settings in which atypical pathogens may be prevalent. In this study, we assessed the ability of our previously developed gene expression classifiers to accurately classify fever associated with ARI in a Sri Lankan cohort.
METHODS
Study Cohort
We collected acute-phase serum, a nasopharyngeal swab, and a peripheral blood sample in PAXgene ribonucleic acid (RNA) tubes (PreAnalytiX, Franklin Lakes, NJ) in consecutive subjects ≥15 years of age hospitalized in the largest tertiary care hospital in southern Sri Lanka from June 2012 to May 2013. Subjects were eligible for enrollment within the first 48 hours of admission if they had documented fever ≥38°C, at least 1 significant respiratory symptom, and lacked signs of a focal infection (eg, urinary tract infection, gastroenteritis). A convalescent serum sample was collected 2–4 weeks after discharge. Demographic and clinical data were obtained at enrollment and during the course of hospitalization. All subjects were noted to be of Sri Lankan descent; specificity of ethnicity was not obtained.
Etiologic Testing and Phenotypic Classification
The nasopharyngeal sample collected from each subject was placed in viral transport media and frozen at −70°C. The sample was tested using the Luminex Integrated System NxTAG Respiratory Pathogen Panel platform (Luminex Corporation, Austin, TX), which detects 19 respiratory viruses and 3 bacteria [27]. Serum specimens were shipped on dry ice for offsite testing. Acute dengue was confirmed using immunoglobulin G enzyme-linked immunosorbent assay (ELISA), virus isolation, real-time reverse-transcription polymerase chain reaction (RT-PCR) for dengue virus (DENV), and RT-PCR for flaviviruses, as previously described [28]. Acute leptospirosis was confirmed as a ≥4-fold rise by microscopic agglutination testing (MAT) with a convalescent titer of ≥200, including in the case of seroconversion, or a single MAT titer of ≥800. We confirmed acute scrub typhus caused by Orientia tsutsumagushi as a ≥4-fold rise in titer from acute to convalescent-phase sample by indirect immunofluorescence assay.
Ribonucleic Acid Sequencing
To assess the performance of our classifiers, we selected a subset of subjects with definitive etiologic diagnoses to move forward for RNA sequencing. Total RNA was extracted using the QIAGEN PAXgene Blood miRNA Kit (QIAGEN, Hilden, Germany). The RNA quantity and quality were assessed using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA), Agilent 2100 Bioanalyzer (Santa Clara, CA), and Qubit 2.0 (Thermo Fisher Scientific) [29].
Ribonucleic acid sequencing was performed in 2 batches. In the first batch, stranded messenger RNA (mRNA) sequencing libraries were prepared using the GLOBINclear Human Kit (Invitrogen, Carlsbad, CA) with a TruSeq Stranded mRNA Library Prep Kit. Libraries were sequenced at 50-base pair (bp) paired-end on an Illumina HiSeq instrument at EA Genomics (Research Triangle Park, NC). Approximately 40 million read pairs per sample were generated. Adapters were removed using Trimmomatic v0.38, reads were aligned to the hg38 reference transcriptome using Bowtie2, and alignments were quantified at the transcript level using Express version 1.5.1. Counts were normalized using trimmed-mean normalization. A sequencing batch effect was corrected with the ComBat method [30].
In the second batch, stranded RNA-Seq libraries were prepared using the commercially available Nugen Universal Plus mRNA-Seq kit (NuGEN Technologies, Redwood City, CA). Nugen’s AnyDeplete-mediated transcript depletion was used to eliminate globin mRNA transcripts from final libraries. Libraries were sequenced at 50-bp paired-end on an Illumina NovaSeq instrument (Duke University Sequencing and Genomic Technologies Core). Approximately 50 million read pairs per sample were generated. Sequence data have been deposited into GEO (accession number GSE149947).
Procalcitonin and C-Reactive Protein Testing
To compare gene expression classifier results to known biomarkers, we performed serum CRP and PCT testing. Quantitative measurement of human CRP by sandwich immunoassay with electrochemiluminescent detection was performed using 2 different platforms depending on the availability and country of location of serum samples. Some serum CRP measurements were performed at 1:1000 dilution, in duplicate, using Human CRP V-PLEX assay kit and QuickPlex SQ 120 Imager (Meso Scale Discovery, Rockville, MD; performed through the Duke Molecular Physiology Institute Biomarkers Core). Other serum CRP measurements were performed using the CRP XL assay kit (DiAgam, Ghislenghien, Belgium; performed through Durdans Laboratory, Sri Lanka).
Quantitative measurement of human PCT was also performed using 2 different platforms depending on the availability and location of serum samples. Some serum PCT measurements were conducted using the VIDAS BRAHMS PCT kit using the ELISA technique via the Mini-VIDAS platform (bioMérieux, Marcy-l’Étoile, France). Other serum PCT measurements were performed using the Exdia PCT (Precision Biosensor Inc., Daejeon, Korea; performed through Durdans Laboratory, Sri Lanka).
Statistical Analysis
The 2 original models included a pan-viral classifier that distinguished 2 classes (viral and nonviral) and an ARI classifier that distinguished 3 classes (separate signatures predicting bacterial infection, viral infection, and noninfectious illness) [25, 26]. Affymetrix probe set identifiers were mapped to RefSeq transcript identifiers: 65 (pan-viral signature), 71 (ARI viral signature), 203 (ARI bacterial signature). The pan-viral model determined classification using a cutoff closest to the top left point of the receiver operating characteristic curve (the “01” method). The ARI classifier used a one-versus-all scheme in which class was assigned by the highest predicted probability. Regularized logistic regression (lasso) was used to predict bacterial and viral infections for both of these prior signatures, and performance characteristics were estimated using repeated, nested 5-fold cross-validation [31, 32]. For the pan-viral classifier, cases were classified as “discordant” if they were misclassified when using the threshold based on the “01” method. For the ARI classifier, cases were classified as “discordant” if the class of infection based on phenotypic classification and the class with the highest probability were different [26].
The area under the receiver operating characteristic (AUROC) curve, sensitivity, and specificity of CRP and PCT for bacterial infections were also determined. Values below the lower limit of detection (LOD) were imputed as half the LOD (1 subject for CRP; 10 subjects for PCT), and subjects at the upper limit of quantitation (LOQ) were assigned the upper LOQ (1 subject for PCT). Cross-validation was used to estimate the AUROC using logistic regression on log-transformed values. All statistical analyses were performed in the R environment for statistical computing (additional packages include ggplot2; OptimalCutpoints) [33].
Ethical Approval
Written informed consent was obtained from all subjects and from parents or guardians of minors, and assent was obtained from all subjects who were 15–17 years of age. The institutional review boards of the Faculty of Medicine, University of Ruhuna (Sri Lanka), Duke University Medical Center (Durham, NC), and Johns Hopkins University (Baltimore, MD) approved this study. The investigators adhered to the policies regarding the protection of human subjects as prescribed by Code of Federal Regulations (CFR) Title 45, Volume 1, Part 46; Title 32, Chapter 1, Part 219; and Title 21, Chapter 1, Part 50 (Protection of Human Subjects).
RESULTS
Phenotypic Classification and Subject Characteristics
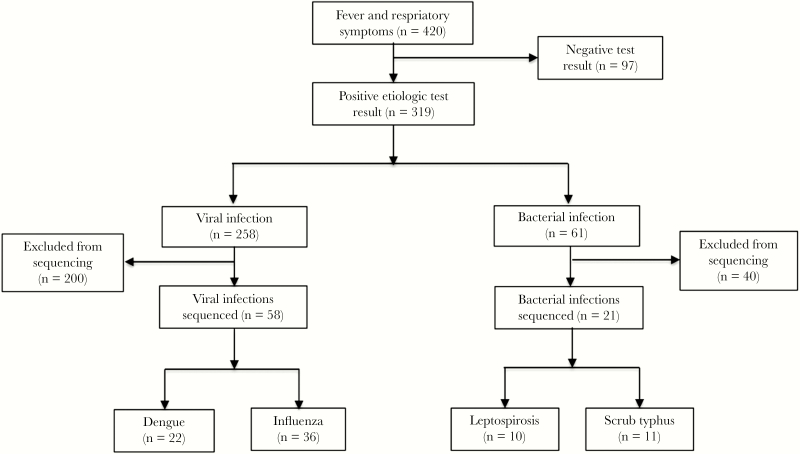
Among 420 subjects with fever and respiratory symptoms, etiologic testing confirmed dengue in 184 (43.8%), influenza in 74 (17.6%), leptospirosis in 35 (8.3%), and scrub typhus in 26 (6.2%). Median age was 32.7 years (interquartile range [IQR], 24.2–47.3), 64.1% were male, and median duration of fever at enrollment was 4 days (IQR, 3–6). Among 319 subjects with a positive etiologic test result, a subset of 79 subjects (24.5% of patients with a positive etiologic test result) was selected at random for RNA sequencing based on funding availability: 58 subjects with viral infections (22 dengue, 36 influenza) and 21 subjects with bacterial infections (10 leptospirosis, 11 scrub typhus) (Figure 1). All subjects were selected for sequencing before any host response information, including CRP or PCT results, were reviewed. All subjects with dengue, leptospirosis, and scrub typhus who were included in the sequencing subset tested negative for respiratory viruses on the Luminex platform.
Figure 1.
Flow diagram showing how subjects were enrolled into the study and selected for ribonucleic acid sequencing in the final analysis set.
Among the 79 subjects for whom RNA sequencing was performed, median age was 37.3 years (IQR, 23.1–52.9) and 54.4% were male (Table 1). Median duration of fever at enrollment was 4 days (IQR, 3–6). Among 5 respiratory symptoms and signs assessed (cough, sore throat, rhinitis/congestion, shortness of breath, pain with breathing), median (IQR) number of respiratory symptoms at enrollment was 2 (1–2) for influenza, 1 (1–2) for dengue, 2 (1–2) for leptospirosis, and 1 (1–2) for scrub typhus. Overall, 56.9% of viral and 61.9% of bacterial infections were treated with antibiotics at enrollment.
Table 1.
Demographic and Clinical Features of Sri Lankan Subjects With Fever and Respiratory Symptoms Who Were Selected for RNA Sequencinga
| Etiologic Classification | ||||
|---|---|---|---|---|
| Viral | Bacterial | |||
| Characteristic | Dengue n = 22 | Influenza n = 36 | Leptospirosis n = 10 | Scrub Typhus n = 11 |
| Age (years) | 31.3 (22.7–51.1) | 37.4 (23.0–57.2) | 40.9 (34.5–43.3) | 37.3 (28.0–50.9) |
| Male | 9 (40.9%) | 22 (61.1%) | 8 (80.0%) | 4 (36.4%) |
| Days of fever at enrollment | 4 (3–6) | 3 (3–4.5) | 4 (2–6) | 7 (4–12) |
| Number respiratory symptoms | 1 (1–2) | 2 (1–2) | 2 (1–2) | 1 (1–2) |
| Rhinitis/congestion | 6 (27.3%) | 15 (42.9%) | 0 | 2 (18.2%) |
| Cough | 11 (50.0%) | 35 (100.0%) | 6 (60.0%) | 9 (81.8%) |
| Sore throat | 4 (18.2%) | 11 (30.6%) | 6 (60.0%) | 2 (18.2%) |
| Shortness of breath | 7 (31.8%) | 6 (16.7%) | 3 (30.0%) | 1 (9.1%) |
| Pain with breathing | 4 (18.2%) | 2 (5.6%) | 4 (40.0%) | 3 (27.3%) |
| Temperature at admission (°F) | 101.5 (99.0–102.0) | 101.4 (99.0–102.0) | 103.2 (102.0–104.0) | 100.0 (98.4–103.0) |
| WBC count × 109/L within 48 hours of admission | 4.0 (2.7–5.2) | 7.4 (6.7–10.2) | 7.8 (5.9–9.6) | 11.3 (5.9–14.0) |
| Platelet count × 109/L within 48 hours of admission | 135 (88–154) | 200 (163–268) | 141 (87–182) | 208 (142–257) |
| Neutrophil% at admission | 71.5 (56.0–80.9) | 74.7 (70.0–78.6) | 80.7 (77.2–86.4) | 76.7 (66.6–82.0) |
| Lymphocyte% at admission | 20.9 (12.6–37.0) | 18.2 (12.8–20.5) | 9.5 (7.2–11.4) | 15.6 (10.8–27.4) |
| Antibiotic use at enrollment | 8 (36.4%) | 25 (69.4%) | 8 (80.0%) | 5 (45.5%) |
| Abnormal chest x-ray (if performed) | 0 of 5 | 1 of 12 (8.3%) | 0 of 3 | 2 of 5 (40.0%) |
Abbreviations: RNA, ribonucleic acid; WBC, white blood cell.
aMedian (25%–75% interquartile range) or frequency (percentage), as appropriate, is listed.
Classifier Performance
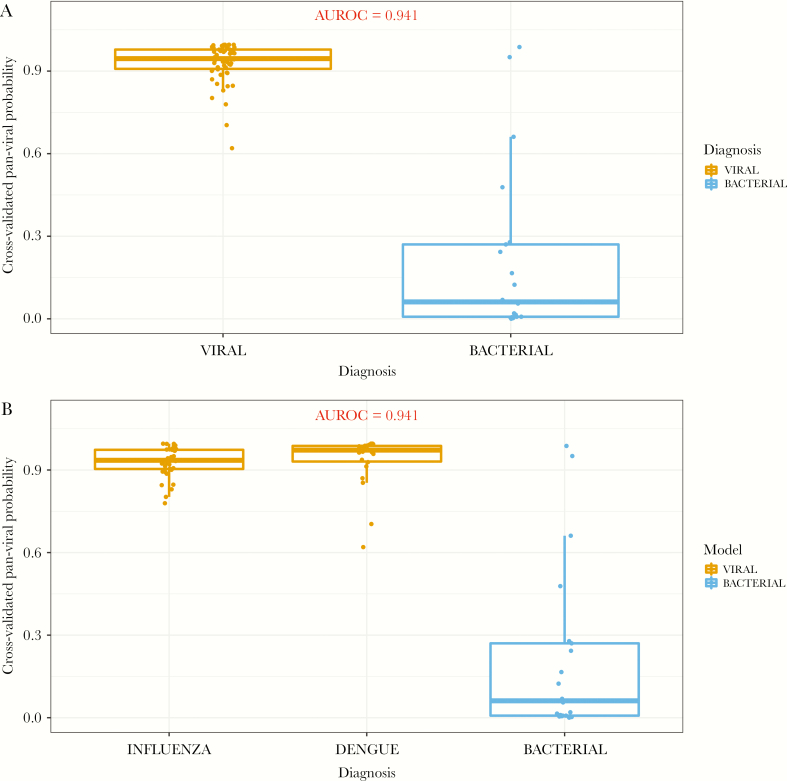
When applying the pan-viral classifier, which was derived to identify subjects with viral infection versus healthy subjects, we observed high predictive accuracy in discriminating viral infections from bacterial infections, with an AUROC of 0.941 (Figure 2a). The 2 viral etiologies (dengue and influenza) had a high probability of being identified by the pan-viral classifier, when compared with the bacterial etiologies (Figure 2b). Sensitivity and specificity of the pan-viral model for identifying viral infections were 98% and 91%, respectively (Table 2). Overall classification accuracy of the pan-viral model was 95%.
Figure 2.
(a) Predicted probabilities when applying the previously developed pan-viral signature in a Sri Lankan cohort of subjects with fever and respiratory symptoms. (b) Predicted probabilities of the individual viral etiologies (dengue and influenza) compared with the nonviral (ie, bacterial) etiologies. AUROC, area under the receiver operating characteristic.
Table 2.
Sensitivity, Specificity, Positive Likelihood Ratio, and Negative Likelihood Ratio of the Pan-Viral Model, Viral and Bacterial Models of the ARI Classifier, CRP, and PCT Among Sri Lankan Subjects
| Model | Sensitivity | Specificity | Positive Likelihood Ratio | Negative Likelihood Ratio |
|---|---|---|---|---|
| Pan-viral | 0.98 | 0.91 | 10.32 | 0.02 |
| ARI classifier, viral model | 0.90 | 0.86 | 6.28 | 0.12 |
| ARI classifier, bacterial model | 0.91 | 0.95 | 17.49 | 0.10 |
| CRP (>10 mg/L) | 1.00 | 0.34 | 1.51 | 0 |
| CRP (>20 mg/L) | 1.00 | 0.50 | 2.00 | 0 |
| PCT (>0.25 ng/mL) | 1.00 | 0.41 | 1.70 | 0 |
| PCT (>0.5 ng/mL) | 0.90 | 0.68 | 2.80 | 0.15 |
Abbreviations: ARI, acute respiratory infection; CRP, C-reactive protein; PCT, procalcitonin.
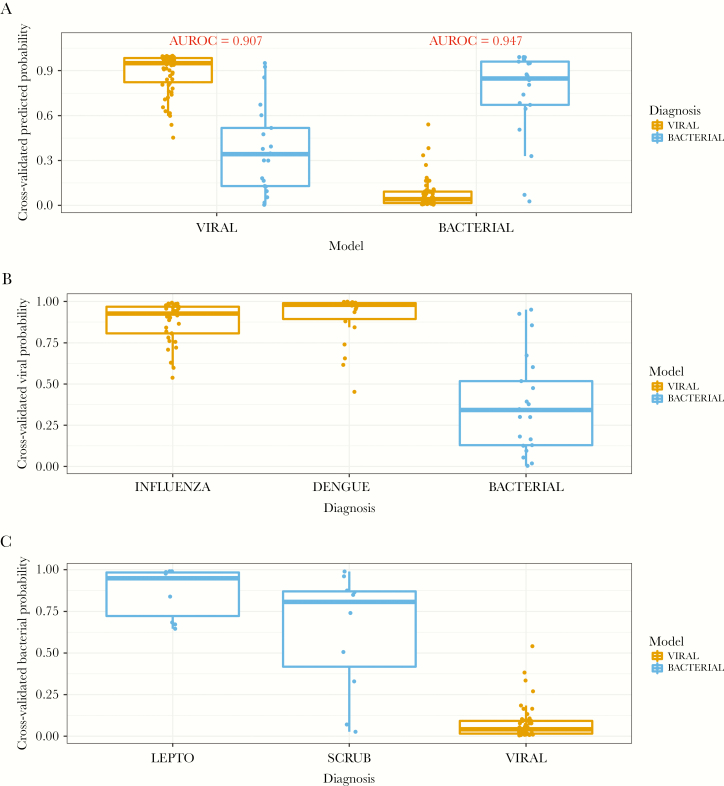
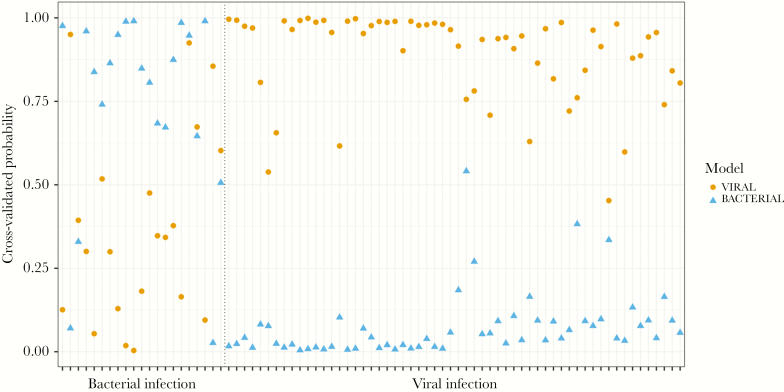
The ARI classifier is composed of multiple signatures that provide independent probabilities of bacterial infection, viral infection, and a noninfectious process. Because our cohort did not include subjects with noninfectious illnesses, we only applied the viral and bacterial models of the ARI classifier, and again we observed high predictive accuracy. For the detection of viral infection, the model had an AUROC of 0.907, which corresponded to 90% sensitivity and 86% specificity (Figure 3a and Table 2). The model also performed well at identifying bacterial infection, with an AUROC of 0.947 corresponding to 91% sensitivity and 95% specificity. The viral model identified the 2 viral etiologies (dengue and influenza) with high probability (Figure 3b), and the bacterial model identified the 2 bacterial infections (leptospirosis and scrub typhus) with high probability (Figure 3c). Figure 4 depicts each subject and predicted probabilities when the bacterial and viral models were applied separately to each individual. Overall classification accuracy of the ARI classifier was 94%.
Figure 3.
(a) Predicted probabilities when applying the viral and bacterial models of the previously developed bacterial/viral/noninfectious acute respiratory infection classifier in a Sri Lankan cohort of subjects. (b) Predicted probabilities of the individual viral infections (dengue and influenza) compared with the bacterial etiologies when applying the viral model. (c) Predicted probabilities of the 2 bacterial infections (leptospirosis and scrub typhus) when applying the bacterial model. AUROC, area under the receiver operating characteristic.
Figure 4.
Predicted probability for each subject when applying the viral and bacterial models of the acute respiratory infection classifier, demarcated by subjects with bacterial infection (left) and viral infection (right). Each subject is represented on the x-axis and has 2 points along the y-axis corresponding to predicted probability when applying the viral model and bacterial model separately.
Discordant Classifications
To better understand discordant classifications, we individually reviewed the cases that were misclassified (Table 3). Overall, there were 7 discordant cases: 4 when using the pan-viral classifier (2 viral and 2 bacterial infection) and 5 when using the ARI classifier (all bacterial infection), with 2 bacterial cases misclassified when using both classifiers (Table 3). The 2 misclassified patients with viral infection had dengue, whereas 4 of 5 misclassified patients with bacterial infection were diagnosed with scrub typhus. For 3 of the 5 misclassified bacterial cases, the bacterial probability was only minimally lower than the viral probability, suggesting coinfection if using independent (not winner-take-all) classifications.
Table 3.
Subjects With Discordant Predictions Compared to Etiologic Testing When Assessing the Pan-Viral Classifier and ARI Classifier, Southern Sri Lanka
| Pan-Viral Classifier | ARI Classifier | |||||||
|---|---|---|---|---|---|---|---|---|
| Confirmed Phenotype | Diagnosis | Probability | Diagnosis | Viral Probability | Bacterial Probability | CRP (mg/L) | PCT (ng/ mL) | Clinical Features |
| Dengue | Bacterial | 0.620 | Viral | 0.453 | 0.335 | .95 | 0.64 | 22 y/o F admitted with 9 d of fever and cough. Clinical diagnosis of dengue at admission and at discharge. No antibiotic therapy at enrollment. WBCa 4.8 (42% neutrophils) and plateletsb 19 000 at admission. CXR clear. No cultures sent. |
| Dengue | Bacterial | 0.704 | Viral | 0.740 | 0.165 | -- | -- | 20 y/o F admitted with 5 d of fever and 1 d of cough. No antibiotic therapy at enrollment. WBC 2.1 (53% neutrophils) and platelets 146 000 at admission. No CXR. No cultures sent. |
| Scrub typhus | Viral | 0.951 | Viral | 0.950 | 0.070 | .25 | 3.36 | 59 y/o F admitted with 16 d of fever and cough. Clinical diagnosis of scrub typhus at admission and lobar pneumonia at discharge. Treated with erythromycin and amoxicillin/ clavulanate at enrollment. WBC 14.0 (84% neutrophils) and platelets 521 000 on d 2 of admission. CXR with alveolar infiltrate in right lower lobe. No cultures sent. |
| Scrub typhus | Viral | 0.987 | Viral | 0.855 | 0.027 | .02 | 1.9 | 48 y/o M admitted with 4 d of fever and 2 d of cough. Clinical diagnosis of dengue vs leptospirosis at admission and dengue at discharge. Treated with 3rd-generation cephalosporin at enrollment. WBC 3.3 (neutrophils 71%) and platelets 17 000 at admission. No CXR. No cultures sent. |
| Scrub typhus | Bacterial | 0.661 | Viral | 0.394 | 0.329 | .11 | 1.24 | 50 y/o M admitted with 14 d of fever and 3 d of cough. Clinical diagnosis of malaria at admission and tuberculosis at discharge. No antibiotic therapy at enrollment. WBC 5.9 (62% neutrophils) and platelets 157 000 on d 2 of admission. No CXR. No cultures sent. |
| Leptospirosis | Bacterial | 0.478 | Viral | 0.673 | 0.646 | .03 | 4.4 | 34 y/o M admitted with 2 d of fever and 1 d of sore throat. Clinical diagnosis of nonspecific viral fever at admission and leptospirosis at discharge. Treated with penicillin at enrollment. WBC 5.2 (77% neutrophils) and platelets 251 000 on d 2 of admission. No CXR. No cultures sent. |
| Scrub typhus | Bacterial | 0.010 | Viral | 0.603 | 0.506 | 151.53 | 26.4 | 27 y/o M admitted with 3 d of fever, rhinitis, and productive cough. Clinical diagnosis of nonspecific viral fever on admission and sepsis at discharge. Treated with amoxicillin/ clavulanate and cloxacillin at enrollment. WBC 23.4 (81% neutrophils) and platelets 302 000 at admission. CXR clear. No cultures sent. |
Abbreviations: ARI, acute respiratory infection; CRP, C-reactive protein; CXR, chest x-ray; d, day; F, female; M, male; PCT, procalcitonin; WBC, white blood cell count; y/o, year old.
aUnits for WBC: ×109/L.
bUnits for platelets: ×109/L.
Biomarker Testing
Of 76 subjects with sera available for both CRP and PCT testing, 56 subjects had viral infection (21 dengue, 35 influenza) and 20 subjects had bacterial infection (9 leptospirosis, 11 scrub typhus). Median CRP was 20.1 mg/L (IQR, 6.58–55.33 mg/L) among subjects with viral infection and 112.10 mg/L (IQR, 81.30–158.67 mg/L) among subjects with bacterial infection. Using a standard CRP cutoff of >10 mg/L, the sensitivity and specificity of CRP for bacterial infections were 100% and 34%, respectively (Table 2) [19]. Specificity increased to 50% when using a higher CRP cutoff of >20 mg/L. The AUROC of CRP for bacterial infections was 0.857. Median PCT was 0.32 ng/mL (IQR, 0.14–0.73 ng/mL) among subjects with viral infection and 1.73 ng/mL (IQR, 1.00–4.09 ng/mL) among subjects with bacterial infection. Using a PCT cutoff of >0.25 ng/mL, which is recommended for respiratory indications, the sensitivity and specificity of PCT for bacterial infection were 100% and 41%, respectively [34]. Specificity increased to 68% when using a higher PCT cutoff of >0.50 ng/mL, which is recommended for sepsis indications. The AUROC of PCT for bacterial infection was 0.829.
DISCUSSION
We demonstrated that ARI gene expression classifiers that were derived in United States and United Kingdom cohorts of primarily European and African descent performed well at identifying bacterial versus viral infection in a South Asian population. Our results suggest that the primary host response to bacterial and viral infection consists of gene pathways that are conserved across race and ethnicity; however, our findings need to be corroborated in other cohorts. In addition, our classifiers had high accuracy regardless of whether the infection was caused by a typical respiratory pathogen, such as influenza, or a pathogen not traditionally associated with ARI, such as O tsutsumagushi. In the case of systemic febrile illnesses such as dengue and leptospirosis, in which nonspecific symptoms prevail and a considerable minority have respiratory symptoms confounding clinical diagnosis, a single class-specific diagnostic may provide great benefit over multiple pathogen-based diagnostics in guiding antimicrobial therapy.
In our Sri Lankan cohort, the ARI classifier had an overall accuracy of 94% at identifying viral and bacterial infection. Although the ARI classifier had slightly lower overall performance compared with the pan-viral classifier, its ability to identify bacterial infection makes it a more advantageous option. In addition, the ARI classifier incorporates a noninfectious classifier as a control group, improving applicability to cohorts with noninfectious causes of respiratory illness. Our classifiers were initially derived in cohorts from the United States and United Kingdom; among those in whom racial information was collected, the majority of these subjects self-identified as being of white or black race. Because differences in biomarker expression between races/ethnicities have been identified in oncologic studies, the finding of similar performance of our classifiers in a racially distinct, South Asian cohort is an important one [35]. The Sri Lankan racial make-up is homogeneous and Asian, with major ethnicities including the Sinhalese (75%), Sri Lankan Tamils (11%), Moors (9%), and Indian Tamils (4%) [36, 37]. Further studies of our classifiers in racially and ethnically diverse populations are warranted.
Others have shown that transcriptional measures of host response are a promising method for distinguishing between bacterial and viral ARI, with genes relevant to the immune response to infection (eg, neutrophil-related versus interferon-related genes) being a distinguishing component of these classifiers [38, 39]. Selected examples of such classifiers include a 10-gene signature for distinguishing between viral and bacterial lower respiratory tract infection and a 35-gene signature for distinguishing between influenza and bacterial infection in pediatric subjects [38, 40]. Our pan-viral classifier consists of 28 unique genes, and the viral and bacterial signatures of our ARI classifier consist of 26 and 71 unique genes, respectively. Two unique genes (LY6E and OASL) are represented in both the pan-viral classifier and viral signature of the ARI classifier, and 3 unique genes (IFI27, IFIT1, and SIGLEC1) are represented in both the pan-viral classifier and the bacterial signature of the ARI classifier. Many of the genes in these classifiers (including the overlap genes of OASL, IFI27, and IFIT1) are involved in potent antiviral responses.
More important, our classifiers performed well despite the bacterial etiologies consisting of atypical, intracellular pathogens such as Leptospira spp and O tsutsumagushi, and the viral etiologies including a nonrespiratory virus (DENV). Nonspecific febrile illnesses may be difficult to diagnose using standard pathogen-based testing in low- or middle-income country (LMIC) settings, and epidemiology may vary widely depending on geographic location and season. In LMICs, erythrocyte sedimentation rate and CRP have classically been used as host-based tests to help identify class of infection, with the latter being more widely used for acute infection. However, both the sensitivity and specificity of CRP are limited at distinguishing bacterial versus viral infections [41]. Procalcitonin has emerged as the most promising single biomarker for distinguishing between bacterial and viral ARI in Western settings. In our study, the sensitivity of both CRP and PCT for bacterial infection was high. Unfortunately, neither test had very high specificity, possibly due to the inclusion of atypical bacterial pathogens. In prior studies in Laos, Cambodia, and Thailand, CRP had better performance characteristics than PCT at distinguishing between viral infections and bacterial infections such as rickettsioses and leptospirosis [19, 42]. In another study of acute febrile illness in Indonesia, CRP outperformed PCT in a cohort that included arboviral infections, murine typhus, leptospirosis, and typical bacterial syndromes such as pneumonia [43]. Gene expression classifiers incorporating multiple mRNA biomarkers into a panel may provide a compelling new diagnostic tool for improving antimicrobial use.
The high cost of gene expression classifier testing will remain a barrier to the practicable application of such technology in LMICs. However, the need for better diagnostics to improve antimicrobial use in LMICs is paramount. Global antimicrobial consumption rose by 65% between 2000 and 2015, with the majority of this increase occurring in LMICs [44]. The associated societal direct cost of antimicrobial resistance is estimated to range from $2 to $20 billion annually in the United States alone; accounting for such societal costs may decrease the acceptable cost threshold for diagnostics, as we have shown in a prior analysis [45–47]. In addition, constant improvements in technology are making measurement of RNA biomarkers more affordable and accessible [48]. To positively impact antimicrobial stewardship, gene expression classifier technology will need to provide a test result in a timely fashion. Our team is currently exploring the translation of this technology to a rapid platform. Analysis of host transcriptomic response to viral infection based on noninvasive samples, such as nasal epithelial cells, also shows promising results and may offer a more practicable approach in the future [49].
We observed few cases in which there was discordance between gene expression classifier performance and etiologic diagnosis. Because reference standard testing was performed, it is possible that initial sample processing or RNA isolation and quantification may have played a role in gene classifier performance. It is interesting to note that, when assessing the ARI classifier, all discordant cases were due to bacterial infections misclassified as viral infections. Because the bacterial etiologies included in this analysis were intracellular, atypical pathogens, they may result in an immune response more characteristic of viral pathogens, as has been shown with intracellular pathogens such as Mycobacterium tuberculosis [50]. In addition, 3 of the 5 misclassified bacterial cases had bacterial probabilities that were only minimally lower than the viral probabilities, suggesting potential coinfection. Finally, 4 of the 5 misclassified subjects with bacterial infection were receiving antibiotics at the time of enrollment, which may have led to decreased performance of the bacterial signature as subjects responded to therapy and immune response lessened.
Some limitations must be noted. We used results from rigorous pathogen-based diagnostic testing among subjects who fit a consistent clinical scenario as our phenotypic gold standard. However, current reference standards for these diagnostics are imperfect. We performed etiologic testing for the most common causes of acute febrile illness in this region of Sri Lanka, but testing was not comprehensive for all known pathogens, and there may have been errors in etiologic classification. The few misclassifications we observed may have been due to an imperfect gold standard or due to true misclassification; as with all diagnostic tests, use of these classifiers would require clinical judgment, and we would expect our host-based diagnostics to be used as a complementary test with pathogen-based diagnostics. We did not have typical bacterial respiratory pathogens such as S pneumoniae in our cohort because we enrolled a cohort presenting with acute febrile illness and significant respiratory symptoms. Our sample size was limited, but we sought to validate a previously developed signature. Our results are also not generalizable to children, because only subjects ≥15 years of age were included. Finally, the performance of our classifiers in the presence of co-infection needs to be evaluated in future studies.
CONCLUSIONS
In conclusion, our pan-viral and ARI gene expression classifiers derived in populations of European and African descent had high accuracy at distinguishing bacterial and viral infection in a South Asian population. Our results must be corroborated in other ARI cohorts of diverse race, ethnicity, and pathogen type to help translate this work to clinical application.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank the subjects, research staff, and laboratory technicians who were involved in this study.
Disclaimer. The views, opinions and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy or decision unless so designated by other documentation.
Financial support. This study was funded by the National Institutes of Allergy and Infectious Diseases (K23AI125677), the US Army Medical Research and Materiel Command under Contract No. W81XWH-16-C-0147, the Office of Naval Research to the Emerging Infectious Diseases Programme, Duke-NUS Graduate Medical School, Singapore, and the Duke Hubert-Yeargan Center for Global Health.
Potential conflicts of interest. M. T. M. reports a patent pending on Biomarkers for the molecular classification of bacterial infection. T. W. B. reports equity in Predigen, Inc. E. L. T. reports grant funding from the Defense Advanced Research Projects Agency (DARPA), the National Institutes of Health/Antibacterial Resistance Leadership Group (ARLG), and the National Institutes of Health/Vaccine and Treatment Evaluation Unit (VTEU); personal fees from bioMerieux; equity in Predigen, Inc.; and patents pending for Biomarkers for the molecular classification of bacterial infection and Methods to diagnose and treat acute respiratory tract infections. G. S. G. reports equity in Predigen, Inc. C. W. W. reports grant funding from DARPA, the National Institutes of Health/ARLG, the National Institutes of Health/VTEU, and Sanofi; equity in Predigen, Inc.; consulting for bioMerieux, Biofire, Giner, and Biomeme; serving on the Advisory Board for Roche Molecular Sciences; and patents pending or issued for Biomarkers for the molecular classification of bacterial infection, Methods to diagnose and treat acute respiratory tract infections, and Methods of identifying infectious disease and assays for identifying infectious disease. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: ID Week, October 2018, San Francisco, CA; American Society of Tropical Medicine and Hygiene Annual Meeting, October 2018, New Orleans, LA.
References
- 1. GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 2017; 17:1133–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shek LP, Lee BW. Epidemiology and seasonality of respiratory tract virus infections in the tropics. Paediatr Respir Rev 2003; 4:105–11. [DOI] [PubMed] [Google Scholar]
- 3. Fleming-Dutra KE, Hersh AL, Shapiro DJ, et al. . Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA 2016; 315:1864–73. [DOI] [PubMed] [Google Scholar]
- 4. Bhavnani D, Phatinawin L, Chantra S, et al. . The influence of rapid influenza diagnostic testing on antibiotic prescribing patterns in rural Thailand. Int J Infect Dis 2007; 11:355–9. [DOI] [PubMed] [Google Scholar]
- 5. Wang J, Wang P, Wang X, et al. . Use and prescription of antibiotics in primary health care settings in China. JAMA Intern Med 2014; 174:1914–20. [DOI] [PubMed] [Google Scholar]
- 6. The Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tillekeratne LG, Bodinayake CK, Simmons R, et al. . Respiratory viral infection: an underappreciated cause of acute febrile illness admissions in southern Sri Lanka. Am J Trop Med Hyg 2019; 100:672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laxminarayan R, Duse A, Wattal C, et al. . Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013; 13:1057–98. [DOI] [PubMed] [Google Scholar]
- 9. The Review on Antimicrobial Resistance. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. 2014. Available at: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. Accessed 1 January, 2020.
- 10. Zaas AK, Garner BH, Tsalik EL, et al. . The current epidemiology and clinical decisions surrounding acute respiratory infections. Trends Mol Med 2014; 20:579–88. [DOI] [PubMed] [Google Scholar]
- 11. Jain S, Self WH, Wunderink RG, et al. ; CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Díaz A, Barria P, Niederman M, et al. . Etiology of community-acquired pneumonia in hospitalized patients in Chile: the increasing prevalence of respiratory viruses among classic pathogens. Chest 2007; 131:779–87. [DOI] [PubMed] [Google Scholar]
- 13. Jain S, Williams DJ, Arnold SR, et al. ; CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015; 372:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodinayake CK, Tillekeratne LG, Nagahawatte A, et al. . Evaluation of the WHO 2009 classification for diagnosis of acute dengue in a large cohort of adults and children in Sri Lanka during a dengue-1 epidemic. PLoS Negl Trop Dis 2018; 12:e0006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trung NV, Hoi LT, Dien VM, et al. . Clinical manifestations and molecular diagnosis of scrub typhus and murine typhus, Vietnam, 2015–2017. Emerg Infect Dis 2019; 25:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith S, Kennedy BJ, Dermedgoglou A, et al. . A simple score to predict severe leptospirosis. PLoS Negl Trop Dis 2019; 13:e0007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crump JA, Gove S, Parry CM. Management of adolescents and adults with febrile illness in resource limited areas. BMJ 2011; 343:d4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsalik EL, McClain M, Zaas AK. Moving toward prime time: host signatures for diagnosis of respiratory infections. J Infect Dis 2015; 212:173–5. [DOI] [PubMed] [Google Scholar]
- 19. Lubell Y, Blacksell SD, Dunachie S, et al. . Performance of C-reactive protein and procalcitonin to distinguish viral from bacterial and malarial causes of fever in Southeast Asia. BMC Infect Dis 2015; 15:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sungurlu S, Balk RA. The role of biomarkers in the diagnosis and management of pneumonia. Clin Chest Med 2018; 39:691–701. [DOI] [PubMed] [Google Scholar]
- 21. Self WH, Balk RA, Grijalva CG, et al. . Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis 2017; 65:183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalil AC, Metersky ML, Klompas M, et al. . Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016; 63:e61–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamat IS, Ramachandran V, Eswaran H, Guffey D, Musher DM. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin Infect Dis 2020; 70:538–42. [DOI] [PubMed] [Google Scholar]
- 24. Zaas AK, Burke T, Chen M, et al. . A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med 2013; 5:203ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zaas AK, Chen M, Varkey J, et al. . Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009; 6:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsalik EL, Henao R, Nichols M, et al. . Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med 2016; 8:322ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen JHK, Lam HY, Yip CCY, et al. . Clinical evaluation of the new high-throughput Luminex NxTAG respiratory pathogen panel assay for multiplex respiratory pathogen detection. J Clin Microbiol 2016; 54:1820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bodinayake CK, Tillekeratne LG, Nagahawatte A, et al. . Emergence of epidemic dengue-1 virus in the southern province of Sri Lanka. PLoS Negl Trop Dis 2016; 10:e0004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 2013; 10:71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118–27. [DOI] [PubMed] [Google Scholar]
- 31. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 32. Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B 1996; 58:267–88. [Google Scholar]
- 33. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 34. bioMerieux. Clinical Guide to Use of Procalcitonin for Diagnosis and Guidance Of Antibiotic Therapy. 2016. Available at: https://www.biomerieux-diagnostics.com/sites/clinic/files/pct_booklet_update_2016-final.pdf. Accessed 15 January, 2019. [Google Scholar]
- 35. Guerrero S, López-Cortés A, Indacochea A, et al. . Analysis of racial/ethnic representation in select basic and applied cancer research studies. Sci Rep 2018; 8:13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Executive Office of the President, Office of Management and Budget Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity. 1997. Available at: https://www.whitehouse.gov/wp-content/uploads/2017/11/Revisions-to-the-Standards-for-the-Classification-of-Federal-Data-on-Race-and-Ethnicity-October30-1997.pdf. Accessed 1 January, 2020.
- 37. Department of Census and Statistics Sri Lanka. Census of population and housing 2012. 2012. Available at: http://www.statistics.gov.lk/PopHouSat/CPH2011/Pages/Activities/Reports/FinalReport/FinalReportE.pdf. Accessed 26 January, 2020.
- 38. Suarez NM, Bunsow E, Falsey AR, et al. . Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis 2015; 212:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parnell GP, McLean AS, Booth DR, et al. . A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 2012; 16:R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramilo O, Allman W, Chung W, et al. . Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007; 109:2066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Díez-Padrisa N, Bassat Q, Machevo S, et al. . Procalcitonin and C-reactive protein for invasive bacterial pneumonia diagnosis among children in Mozambique, a malaria-endemic area. PLoS One 2010; 5:e13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wangrangsimakul T, Althaus T, Mukaka M, et al. . Causes of acute undifferentiated fever and the utility of biomarkers in Chiangrai, northern Thailand. PLoS Negl Trop Dis 2018; 12:e0006477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prodjosoewojo S, Riswari SF, Djauhari H, et al. . A novel diagnostic algorithm equipped on an automated hematology analyzer to differentiate between common causes of febrile illness in Southeast Asia. PLoS Negl Trop Dis 2019; 13:e0007183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klein EY, Van Boeckel TP, Martinez EM, et al. . Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A 2018; 115:E3463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thorpe KE, Joski P, Johnston KJ. Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff (Millwood) 2018; 37:662–9. [DOI] [PubMed] [Google Scholar]
- 46. Roberts RR, Hota B, Ahmad I, et al. . Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis 2009; 49:1175–84. [DOI] [PubMed] [Google Scholar]
- 47. Tillekeratne LG, Bodinayake C, Nagahawatte A, et al. . Use of clinical algorithms and rapid influenza testing to manage influenza-like illness: a cost-effectiveness analysis in Sri Lanka. BMJ Glob Health 2019; 4:e001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet 2019; 20:631–56. [DOI] [PubMed] [Google Scholar]
- 49. Yu J, Peterson DR, Baran AM, et al. . Host gene expression in nose and blood for the diagnosis of viral respiratory infection. J Infect Dis 2019; 219:1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berry MP, Graham CM, McNab FW, et al. . An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.