Fig. 3. Sample-specific matrix effect and a linear response observed with optimized FI-MS.
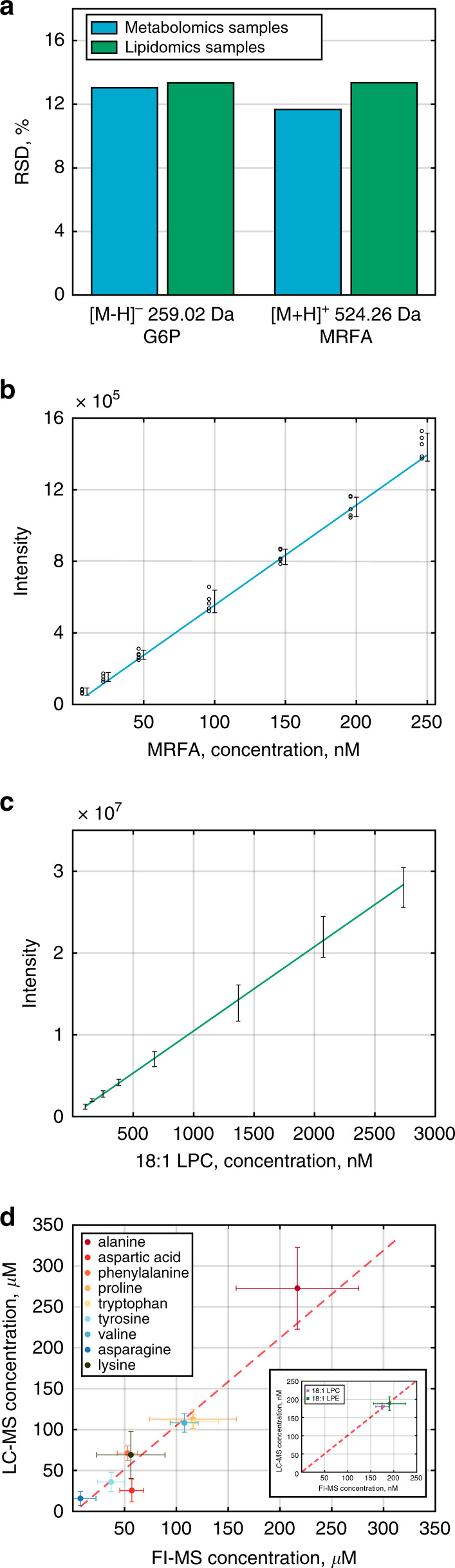
a Applying FI-MS method with 8 optimized ranges in negative ionization mode for metabolomics and lipidomics analysis of serum samples from 20 donors and adding 250 nM of glucose 6-phosphate (G6P); and in positive ionization mode, adding 150 nM of Met-Arg-Phe-Ala tetrapeptide (MRFA); y axis represents the RSD% of the measured intensity of these compounds across the 20 analyzed samples. Applying FI-MS with eight optimized ranges in positive ionization mode for metabolomics analysis of serum samples, adding increasing concentrations of taurocholic acid (b) and lipidomics analysis in positive ionization mode adding increasing concentrations of 18:1 lyso phosphocholine (LPC) (c), demonstrating a linear response in both cases. Data are mean values ± SD, n = 5 (10 for LPC measurement) independent repetitions of the FI-MS analysis. d Correlation between FI-MS and LC-MS-based concentration measurements of nine amino acids and two lipids in serum. Error bars show 95% confidence intervals. Source data are provided as a Source data file.
