Fig. 3.
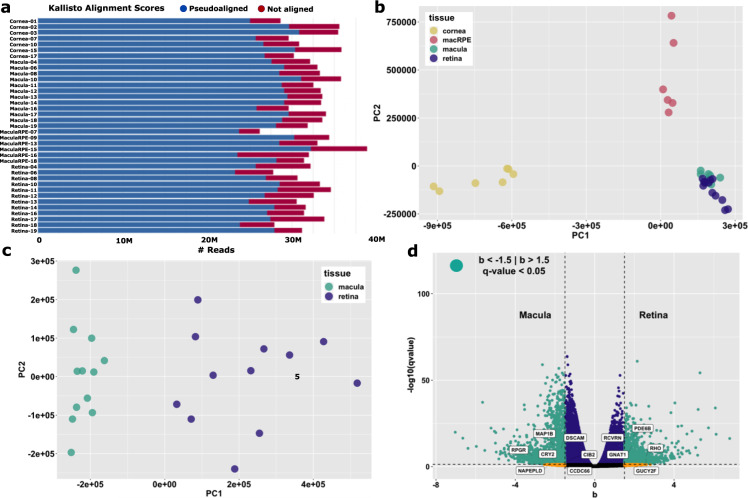
Quality assessment of read alignment and sample variance. The majority of high quality sequence reads successfully aligned to the human hg38 reference transcriptome using the Kallisto pseudoaligner (a). Kallisto transcript quantification of each sample was used for two dimensional principal component analysis (PCA) of sample variance with principle component 1 (PC1) and principle component 2 (PC2) accounting for the majority of each sample’s variance. PCA was used to visualize variance between the 4 distinct sample groups as well as similarity within sample replicates for all 37 samples (b). PCA was separately applied to 24 tightly clustered central retina and peripheral retina samples (c). Differentially expressed transcripts between central and peripheral retina samples plotted based on q-value [-log10(qval)] and fold change [b] demonstrates RHO, PDE6B, RCVRN, GUCY2F, and GNAT1 are preferentially expressed in the peripheral retina whereas CIB2, MAP1B, CRY2, NAPEPLD, DSCAM, CCDC66, and RPGR are representative transcripts preferentially expressed in the central retina (d).