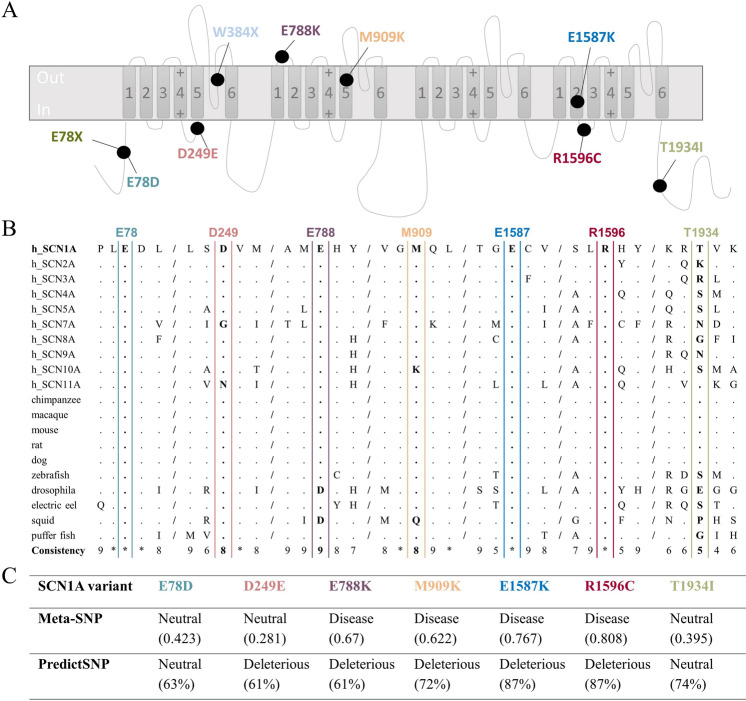
Figure 3.
Overview of the studied mutants – localisation, evolution conservation, variant prediction. (A) Topology diagram of SCN1A variants. Schematic representation of the sodium channel molecular complex illustrating the location and amino acid substitution of the studied SCN1A variants. (B) The amino acid sequence alignment of the Nav α-subunit family, showing the evolutionary conservation of the residues (boxed). E78, D249, E788, M909, E1587, R1596, and T1934 are shown in bold. Residues that are identical to the SCN1A sequence are displayed as dots. The conservation scoring was performed by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 for the most conserved alignment position. Conservation in all studied sequences is marked as *. (C) The results of SCN1A variant prediction by Meta-SNP and PredictSNP. “Neutral” referred to a neutral variant, “Disease” and “Deleterious” to disease-causing variants. The variant was predicted to be disease-causing when Meta-SNP predictions were >0.5. The percentages are the expected prediction accuracies by PredictSNP.