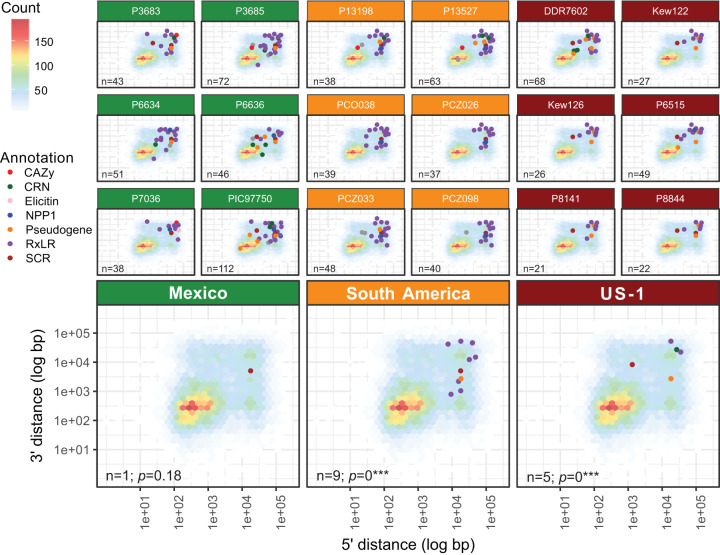
FIG 3.
Gene loss in pathogenicity factors and pseudogenes occurs in individuals within populations regardless of clonal or sexual reproductive mode. The top three rows show results for individual samples, and the bottom row shows the summary for the populations sampled from Mexico (green), South America (orange), and the US-1 clonal lineage (red). Gene loss is common in individuals that may lose between 21 and 112 pathogenicity genes or pseudogenes. However, when comparing multiple samples within a population, few losses are shared. Gene loss is happening on an individual basis and is not dependent on ancestry. This pattern is observed in sexually reproducing populations (Mexico) as well as in clonally reproducing populations (South America and US-1). Gene loss is defined here as a gene from a sample with an adjusted average read depth of at least 12 and where 0 positions (base pairs) in the gene were sequenced (BOC = 0). The background for each panel is a heatmap indicating gene abundance in relation to their 5′ and 3′ intergenic spacing. Points indicate gene deletions and are colored by their functional annotation as defined in Haas et al. (23).