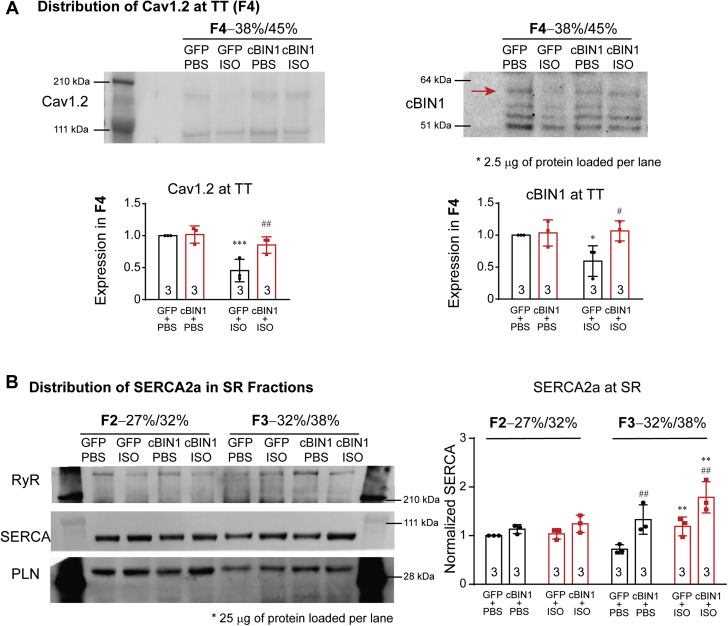
Figure 5.
Sucrose Gradient Fractionation of Cardiac Microsomes
(A) Representative Western blots of Cav1.2 and cBIN1 in the F4 (TT) fraction of cardiac microsome from GFP+PBS, GFP+ISO, cBIN1+PBS, and cBIN1+ISO hearts (2.5 μg proteins loaded per lane). Quantification is included in the bar graphs (n = 3 hearts per group). (B) Representative Western blots of RyR, PLN, and SERCA2a in the F2 (longitudinal SR enriched) and F3 (jSR enriched) fractions of cardiac microsome from GFP+PBS, GFP+ISO, cBIN1+PBS, and cBIN1+ISO hearts (25 μg protein loaded per lane). Quantification of SERCA2a in F2 and F3 is included in the bar graphs to the right (n = 3 hearts per group). Data are expressed as mean ± SD. Two-way ANOVA was used followed by Fisher LSD test for multiple comparison. ∗p < 0.05, ∗∗p <0.01, and ∗∗∗p < 0.001 for PBS vs. ISO comparison within each AAV9 treatment group; #p < 0.05 and ##p < 0.01 for GFP vs. cBIN1 comparison within each drug infusion group. jSR = junctional sarcoplasmic reticulum; PLN = phospholamban; RyR = ryanodine receptor; SD = standard deviation; SERCA2a = sarcoplasmic reticulum calcium ATPase pump 2a; other abbreviations as in Figure 1.