Figure 1. Two major pathways of DNA double strand break repair.
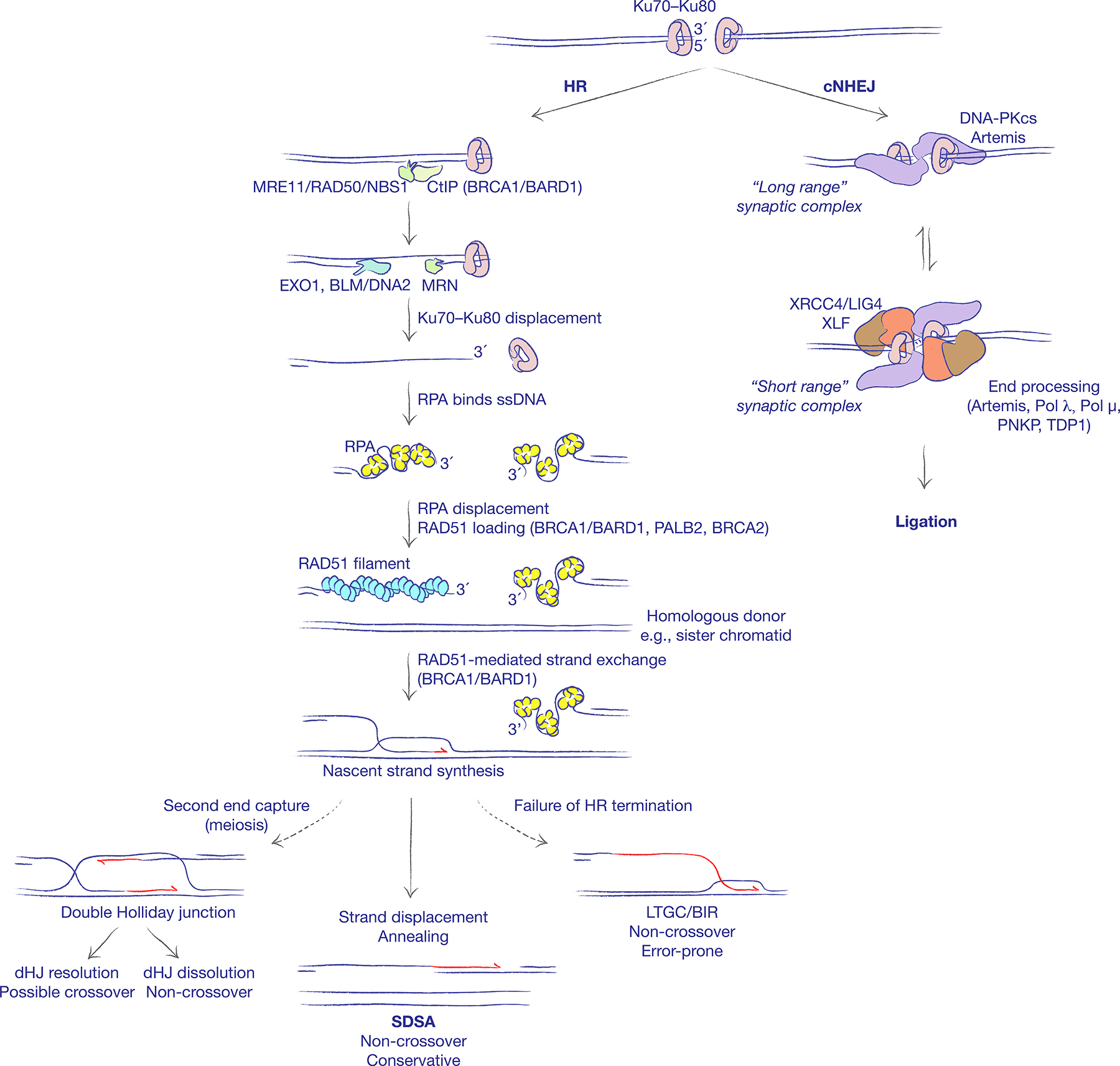
The binding of the Ku70–Ku80 heterodimer to DNA ends schedules repair of DNA double strand breaks (DSBs) by classical non-homologous end joining (cNHEJ). cNHEJ entails formation of a ‘long range’ synaptic complex, which is in equilibrium with a ‘short range’ synaptic complex. End processing by cNHEJ enzymes (as shown) and ligation are restricted to the short range complex. PNKP: Polynucleotide kinase-phosphatase. TDP1: Tyrosyl-DNA phosphodiesterase 1. The default engagement of cNHEJ can be disrupted by DNA end resection. The nuclease activity of MRE11 converts the blunt end into a 3ʹ single-stranded DNA (ssDNA) tail, displacing Ku70–Ku80 from the DNA end and establishing the possibility of repair by homologous recombination (HR). The replication protein A (RPA) complex avidly binds to ssDNA and must be displaced by recombination mediators to enable the formation of a RAD51 nucleoprotein filament. BRCA2 is the major recombination mediator in mammalian cells, likely acting in concert with PALB2 and the BRCA1–BARD1 heterodimer. Interactions between the two DNA ends at the recombination synapse, and operations on the D-loop formed following synapsis, influence which HR sub-pathway is engaged. The non-crossover synthesis-dependent strand annealing (SDSA) pathway is the predominant repair pathway in somatic cells. In meiotic cells, formation of a double Holliday junction (dHJ) intermediate can lead to crossing over. A failure to engage the second end of the break, or failure to displace the nascent strand leads to aberrant replicative HR responses of long tract gene conversion (LTGC) and break-induced replication (BIR). Established roles for BRCA gene products in HR are indicated in parentheses.
