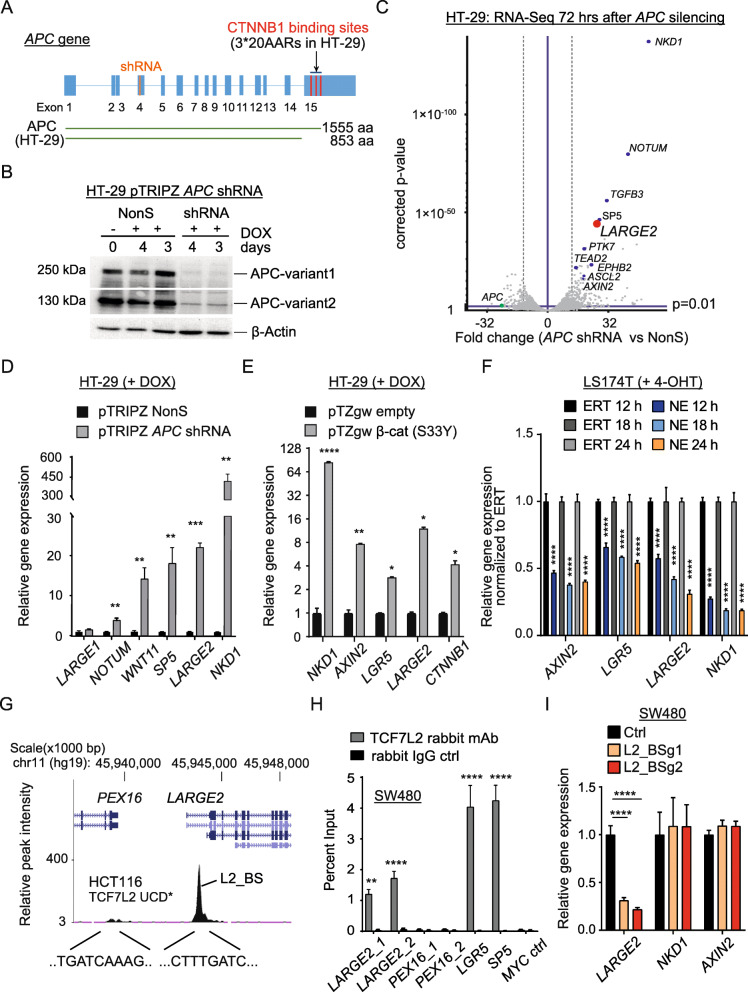
Fig. 1.
LARGE2 is a direct target of Wnt signaling in colorectal cancer. A)APC gene structure, exons indicated as blue bars. A short hairpin RNA (shRNA, indicated in orange) was used to silence APC. Truncated APC variants in HT-29 CRC cells are shown as green lines. aa: amino acids. B) Immunoblot analysis of APC and β-actin on WCL from HT-29 cells treated with 500 ng/ml DOX to induce expression of APC-targeting shRNA or a non-silencing shRNA (NonS). C) Volcano plot showing genes deregulated 72 h after silencing of APC in HT-29 cells, as analyzed via RNA-Seq on biological duplicates. Cut off: normalized p-value < 0.01 (horizontal blue line), fold change > 4 (vertical dotted lines). D,E) qRT-PCR analysis of indicated genes up-regulated upon conditional APC silencing (D) or expression of S33Y-mutated CTNNB1 (E) in HT-29 cells for 72 h (+ 500 ng/ml DOX). Results are shown as mean ± SD (n = 3); * p < 0.05; ** p < 0.01; **** p < 0.0001. F) qRT-PCR analysis of indicated genes in LS174T-NE or -E cells after treatment with 400 nM 4-OHT for the indicated times. Results are shown as mean ± SD (n = 3); **** p < 0.0001. G) Structure of the proximal LARGE2 genomic locus (UCSC Browser) showing a potential TCF7L2 binding site (L2_BS) in the first LARGE2 intron. * Source: HCT-116 TCF7L2 UC Davis ChIP-seq Signal from ENCODE/SYDH (Peggy Farnham lab). H) qChIP analysis on genomic DNA from SW480 cells. The amount of DNA immunoprecipitated with TCF7L2 antibody or rabbit IgG-control in each sample is shown as percentage of chromatin input. Results are shown as mean ± SD (n = 3); ** p < 0.01; **** p < 0.0001. I) qRT-PCR analysis of indicated genes in stably transduced SW480 cells upon CRISPR/eCas9-mediated targeting of the TCF7L2_BS in the first intron of LARGE2 via two different guide RNAs. Results are shown as mean ± SD (n = 5); **** p < 0.0001