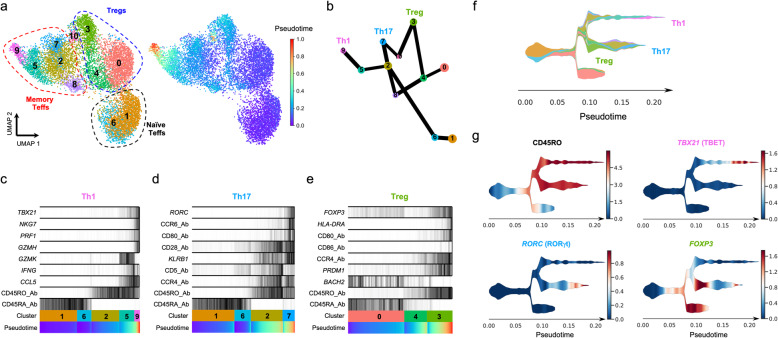
Fig. 3.
Pseudotime analysis reveals distinct trajectories of CD4+ T cell differentiation in vivo. a UMAP plots depicting the inferred diffusion pseudotime of each single cell in the identified T cell clusters. b Graph reconstructing the developmental trajectories between the identified T cell clusters. Edge weights represent confidence in the presence of connections between clusters. The analysis was performed in the combined transcriptional and proteomics data using the partition-based graph abstraction (PAGA) method. c–e Reconstructed PAGA paths for the differentiation of the identified Th1 (c), Th17 (d) and Treg (e) lineages. Expression of the lineage-specific transcription factors and selected differentially expressed genes is depicted for each trajectory. f Schematic representation of the identified lineage differentiation trajectories using the single-cell trajectories reconstruction (STREAM) method. Colour code corresponds to the cluster assignment depicted in a. g Expression of the memory-associated CD45RO isoform and the lineage-specific transcription factors TBX21 (encoding TBET), RORC (encoding RORγt) and FOXP3 is depicted along the identified developmental branches