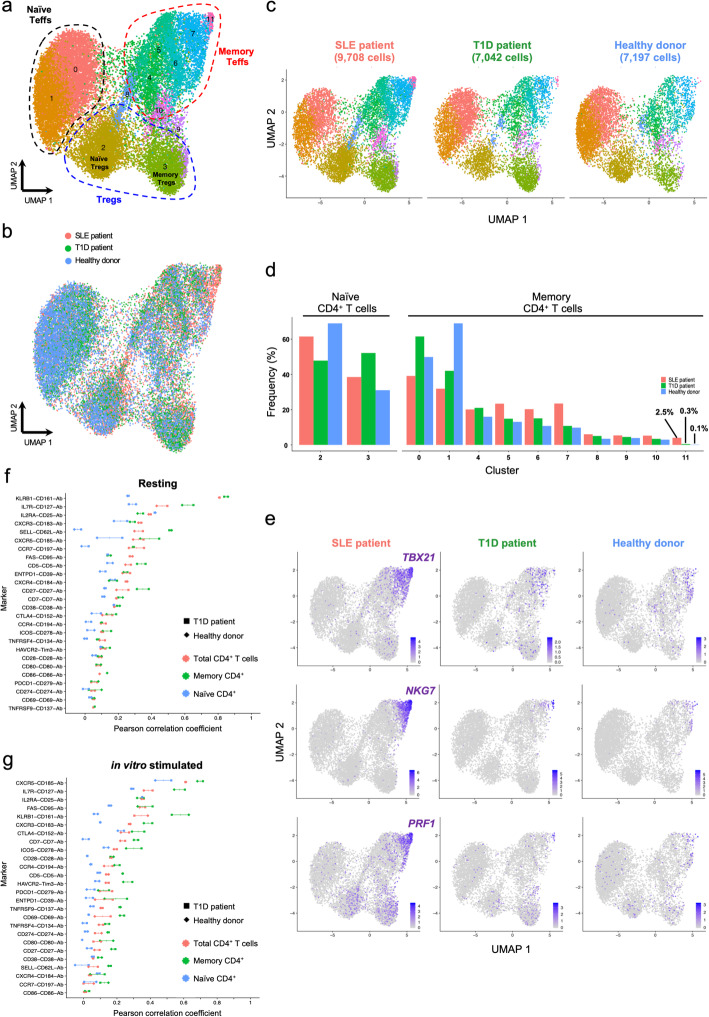
Fig. 7.
Data from independent experiments can be robustly integrated. a UMAP plot depicting the clustering of resting primary CD4+ T cells from one systemic lupus erythematosus (SLE; n = 9708 cells) patient, one type 1 diabetes (T1D; n = 7042 cells) patient and one healthy donor (n = 7197 cells). Data were integrated from two independent experiments using the same CD4+ T cell FACS sorting strategy (described in Fig. 1a). b Alignment of the integrated targeted transcriptomics and proteomics data generated from the three assessed donors in two independent experiments. c UMAP plots depicting the donor-specific clustering of the CD4+ T cells. d Relative proportion of the identified CD4+ T cell clusters in each donor. Frequencies were normalised to either the annotated naive or memory compartments to ensure higher functional uniformity of the assessed T cell subsets and to avoid alterations associated with the declining frequency of naive cells with age. e UMAP plots depicting the relative expression of the canonical Th1 transcription factor TBX21 (encoding TBET) and the effector cytokines NKG7 and PRF1 on the three assessed donors. f, g Correlation (Pearson correlation coefficient) between mRNA and protein expression for 26 markers with concurrent mRNA and protein expression data in resting (f) and in vitro-stimulated (g) CD4+ T cells. The correlation was calculated in the total CD4+ T cells (red) or in the CD45RA− memory (green) or CD45RA+ naive (blue) T cell subsets separately. Individual-level correlation in the type 1 diabetes (T1D) patient (square) and healthy donor (diamond) and median correlation in both donors are displayed in the figure