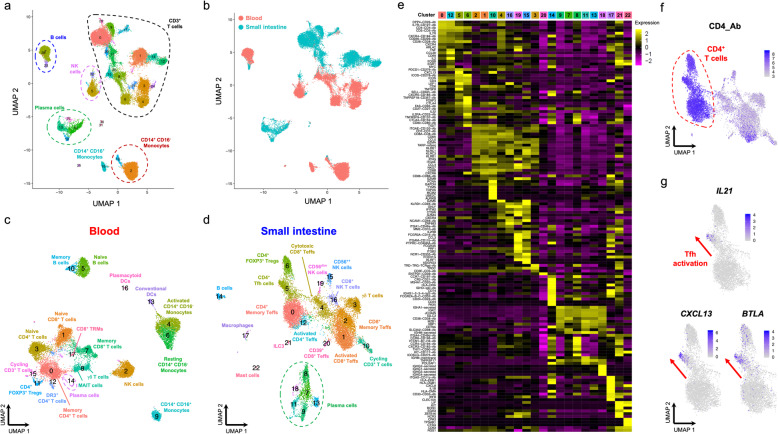
Fig. 8.
Targeted scRNA-seq and proteomics approach delineates distinct functional subsets in a heterogeneous population of CD45+ immune cells isolated from blood and tissue. a UMAP plot depicting the clustering of the targeted scRNA-seq and transcriptional data of a heterogeneous population of total CD45+ cells (n = 31,907) isolated from blood and a paired duodenal biopsy from two coeliac disease (CD) patients with active disease. b Sample Tag information identifies samples isolated from the blood (red) or from the paired duodenal biopsy (teal). c, d UMAP plot depicting the clustering of the CD45+ cells isolated from the blood (c) or the paired duodenal biopsy (d). e Heatmap displaying the top 10 differentially expressed genes in each identified cluster from the CD45+ immune cells isolated from the duodenal biopsies. f UMAP plot depicting the expression of CD4 at the protein level (AbSeq) within the CD3+ T cells isolated from the small intestine. g Gradient of expression of the Tfh effector genes IL21, CXCL13 and BTLA in tissue-resident CD4+ T cells. DR3, death-receptor 3 (encoded by TNFRSF25); TRM, tissue-resident memory T cells; MAIT, mucosal-associated invariant T cells; ILC3, type 3 innate lymphoid cell