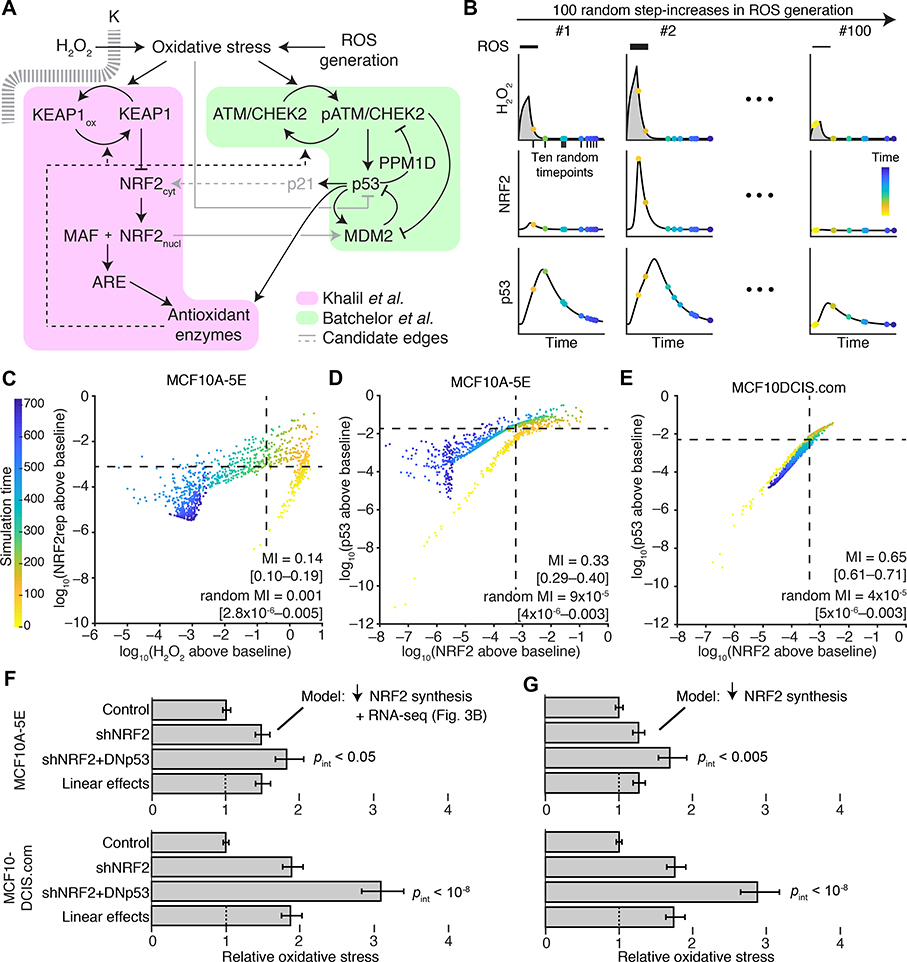
Fig. 5. NRF2–p53 pathway coordination and synergistic phenotypes are captured by an integrated-systems model of oxidative stress.
(A) Connecting NRF2 and p53 signaling models (68, 69, 71) through oxidative-stress activators and antioxidant target enzymes. Additional crosstalk linking oxidative stress to p53 inhibition (73), p53 to NRF2 through p21 (76), and NRF2 to MDM2 (74, 75) was considered (gray). (B) Simulation strategy for quantifying association between signaling intermediates. The model was challenged with various ROS production rates and randomly sampled at multiple intermediate time points (yellow to blue). Integrated intracellular H2O2 (gray) is used for phenotype predictions related to NRF2 and p53 perturbation. (C) Intracellular H2O2 concentration is associated with a reporter of NRF2 stabilization (NRF2rep) following simulated step increases in ROS production rate as illustrated in (B). (D and E) Coordination of NRF2 and p53 stabilization in the oxidative-stress model and in simulations of premalignancy through the computational approach illustrated in (B). (F and G) Modeling NRF2 knockdown by reduced synthesis captures the synergistic oxidative-stress profile of cells harboring dual perturbation of the NRF2 and p53 pathways. In (F), transcriptional changes secondary to NRF2 knockdown were added to the model according to the results in Fig. 3B. For (C) to (E), simulated time points are log-scaled and background-subtracted mutual information (MI) with 90% CI for ten time points from n = 100 random ROS generation rates. For (F) and (G), time-integrated intracellular H2O2 profiles are scaled to the unperturbed simulations and reported as the mean oxidative stress with 90% CI from n = 100 random ROS generation rates. Statistical interaction between shNRF2 and DNp53 perturbations (pint) was assessed by two-way ANOVA with replication.