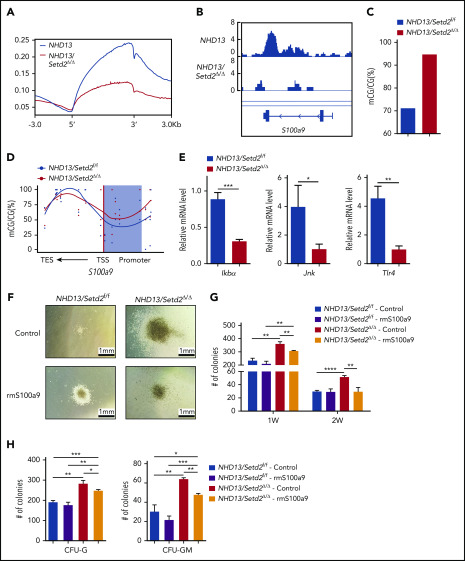
Figure 7.
S100a9 is directly regulated by Setd2, and restoration of S100a9 abrogates the effect of Setd2 deficiency on the NHD13 HSPCs. (A) A profile of H3K36me3 across the gene body and the upstream (5′) and downstream (3′) regions. ChIP-seq analyses were performed with spleen c-Kit+ cells isolated from the NHD13 and NHD13/Setd2Δ/Δ mice at leukemia stage. (B) A decrease occurred in H3K36me3 in the S100a9 gene locus of the NHD13/Setd2Δ/Δ cells relative to that in the NHD13 cells. (C-D) Different DNA methylation in the S100a9 promoter (C) and a profile of DNA methylation across the S100a9 gene locus (D) in the NHD13 and NHD13/Setd2Δ/Δ cells. TSS, transcription start sites; TES, transcription end sites; the shading denotes the promoter region. (E) qPCR analysis of Ikba, Jnk, and Tlr4 in the NHD13 and NHD13/Setd2Δ/Δ cells. (F) Micrographs of representative colonies in the CFU assays with the NHD13 and NHD13/Setd2Δ/Δ HSPCs. Cells (3 × 103) were plated for each assay. Recombinant mouse S100a9 protein (rmS100a9) was added at a final concentration of 4 μg/mL. (G-H) The number of indicated colonies in the CFU assays. CFU-G and CFU-GM were counted 1 week after plating. *P < .05; **P < .01; ***P < .001; ****P < .0001.