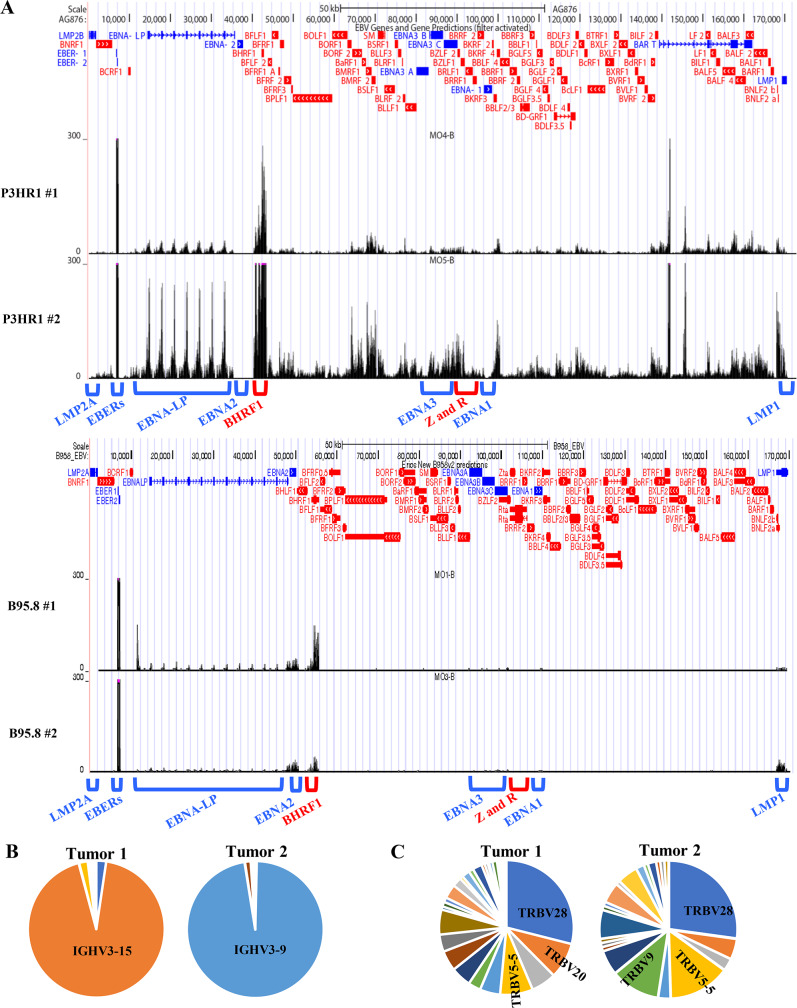
Fig 9. P3HR1 virus-infected tumors express a complex mix of latent and lytic genes and are oligoclonal.
A. RNA-seq reads were mapped to the EBV genomes of 2 different tumors infected with P3HR1 virus (top panel), or two different tumors infected with B95.8 virus (middle panel). The locations of the Cp and the EBER, EBNA-LP, EBNA2, BHRF1, EBNA3, BZLF1 (Z), BRLF1 (R), EBNA1, LMP1 and LMP2A transcripts are indicated above the EBV genome map. P3HR1 “1” tumor is derived from mouse #6, and P3HR1 “2” tumor derived from mouse #7 in S1 Table. B. RNA-seq analysis was performed using RNA isolated from 2 different P3HR1 infected tumors shown in top panel in A. The relative frequency of the IGH transcripts containing various different IGHV genes is shown for each tumor. C. The frequency of the T cell receptor TRBV gene reads in the RNA-seq analysis of each P3HR1 tumor was determined by comparing the number of reads from each individual TRBV gene to the total number of TRBV gene reads.