Figure 7. Model of Epigenetic Changes at Pluripotency Gene Enhancers during Stem Cell Differentiation.
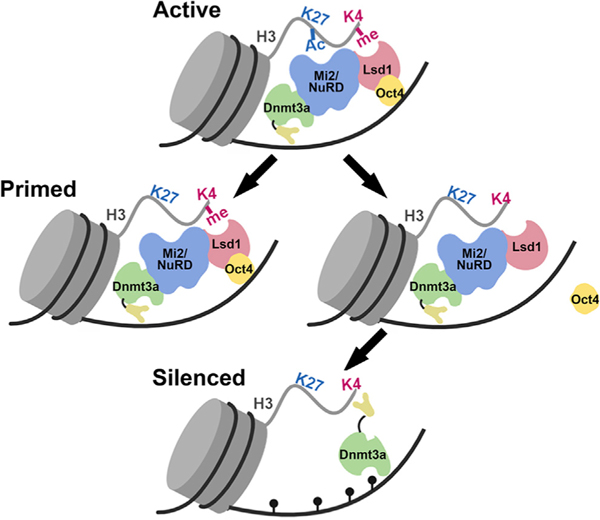
</p>In an undifferentiated state, the pluripotency gene enhancers (PpGe) are active, bound by the coactivator complex, and contain chromatin modifications, including H3K4m2/1 and H3K27Ac. In response to the signal of differentiation, the dissociation of the coactivator complex, including Oct4, is followed by the activity of the Lsd1-Mi2/NuRD complex, which facilitates enhancer silencing. The histone deacetylase (HDAC) removes H3K27Ac at PpGe, and Lsd1 demethylates H3K4me1, followed by DNA methylation by Dnmt3a. However, in F9 ECCs, Lsd1 activity is inhibited in the presence of Oct4, causing retention of H3K4me1. The ADD domain of Dnmt3a cannot interact with the H3K4 methylated histone tail and will potentially remain in the autoinhibited state, thus preventing DNA methylation at these sites. Consequently, PpGe, instead of being silenced, acquire a “primed” state. Black pins represent methylated CpGs.
