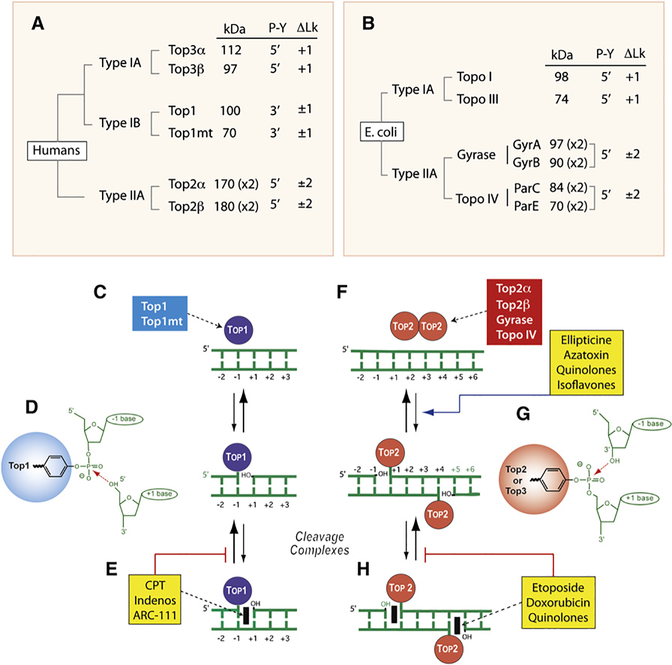
Figure 1. Overview of DNA Topoisomerases.
(A) Classification of human DNA topoisomerases. kDa: molecular masses calculated from polypeptide composition. (x2): dimer. Type IB are the only enzymes that form cleavage complexes (cc) with 3′-phosphotyrosyl (3′-P-Y) intermediates. ΔLk: linking number change produced by each catalytic cycle. Type I enzymes change Lk in steps of one and type II in steps of two as they cleave one and both strands of DNA, respectively. Type IA enzymes are the only enzymes that relax only negative but not positive supercoiling (ΔLk = +1).
(B) Classification of E. coli DNA topoisomerases.
(C) Noncovalent binding of type IB enzymes.
(D) Scheme of the 3′-phosphotyrosine covalent bond in the Top1cc. The arrow indicates the reversible (religation) reaction, which is favored under normal conditions.
(E) Trapping of the cleavage complex by camptothecin (CPT) and the Top1 inhibitors (see Figure 3).
(F) Noncovalent binding of type IIA enzyme homodimers (A2B2 and C2E2 in the case of gyrase and Topo IV, respectively).
(G) Scheme of the 5′-phosphotyrosine covalent bond in the Top2cc. The arrow indicates the reversible (religation) reaction, which is favored under normal conditions. Note the four base pair stagger with 5′-overhang on opposite strands characteristic of all type IIA enzymes.
(H) Trapping of the cleavage complex by etoposide, doxorubicin, or quinolones (see Figure 3). Note that some Top2 poisons increase the steady-state levels of Top2cc by increasing cleavage (ellipticines, azatoxin, quinolones, and isoflavones).