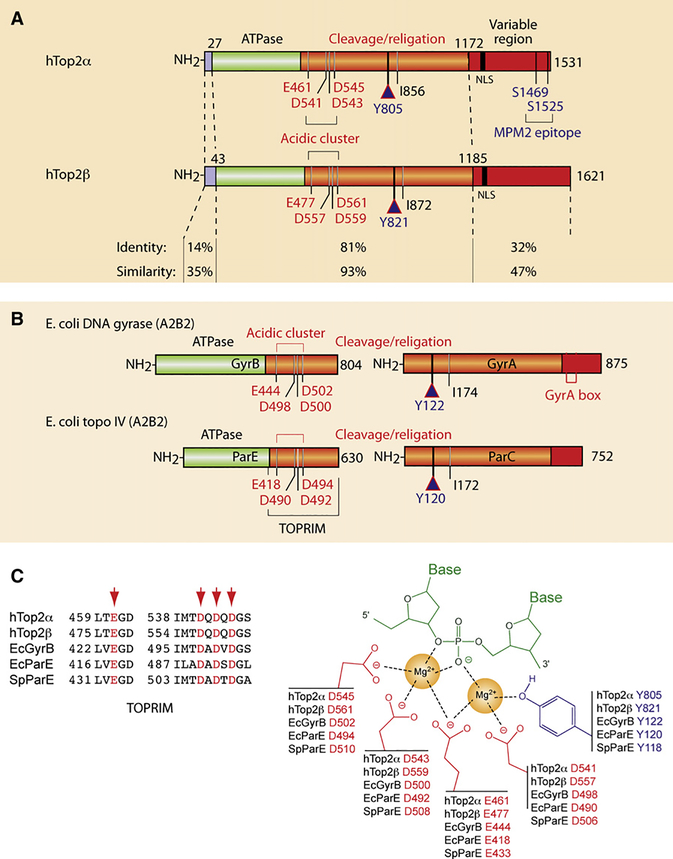
Figure 4. Structural Overview of the Type IIA Topoisomerases Targeted by Anticancer and Antibacterial Drugs.
(A) Schematic representation of the human Top2 enzymes Top2α and β. Note the high similarity of their ATPase and cleavage/religation domains, whereas their N- and C-terminal regions are more divergent. Conserved catalytic residues are indicated including the catalytic tyrosines Y805 and Y821 in Top2α and β, respectively.
(B) Schematic representation of the bacterial Top2 enzymes gyrase and Topo IV. Gyrase is a heterotetramer encoded by two genes: GyrA and GyrB (A2B2; see Figure 1B). Topo IV is also a heterotetramer encoded by two genes: ParC and ParE (C2E2; see Figure 1B). Conserved catalytic residues are indicated including the catalytic tyrosines Y122 andY120 ingyrase and Topo IV, respectively.
(C) Conserved interaction of type IIA enzymes with DNA by their TOPRIM motifs (alignment at left). The right scheme represents the two-metal-binding model between the TOPRIM motif and DNA (modified from Deweese et al., 2008).