Fig. 7.
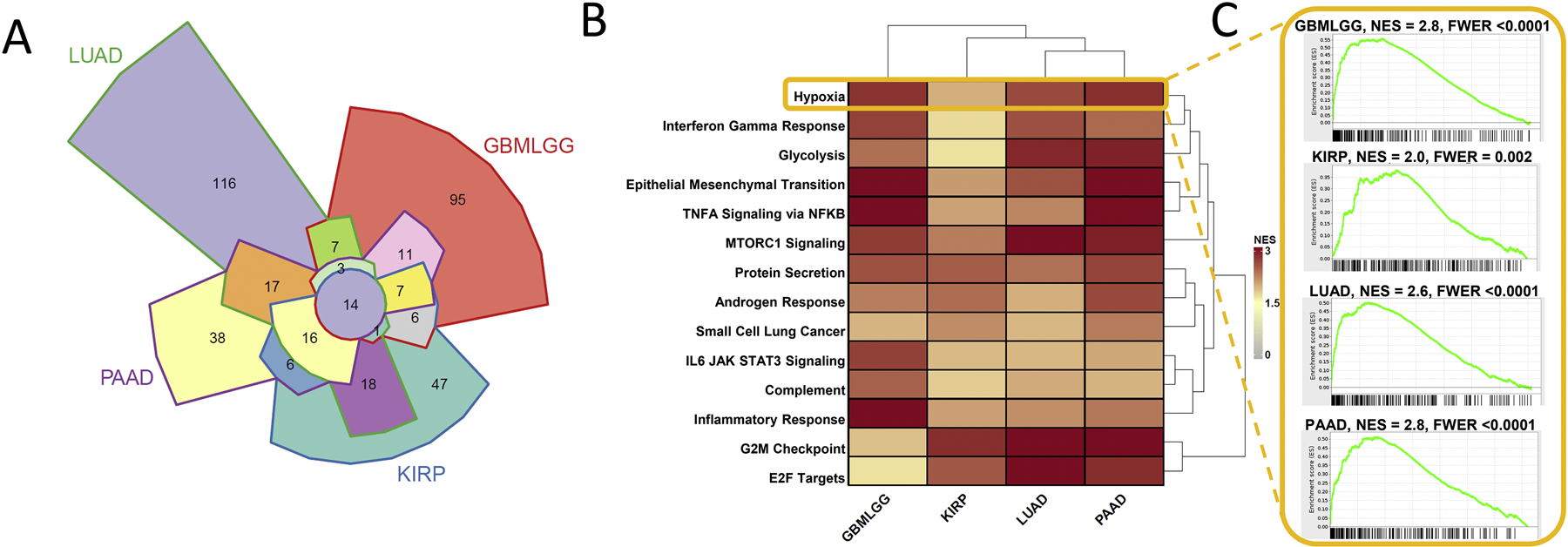
Common gene sets enriched for genes that are co-expressed with ERO1A. A) Gene set enrichment analysis (GSEA) was used to identify enriched pathways with genes that are co-expressed with ERO1A. A Chow-Ruskey diagram shows overlap of significant gene sets correlated with ERO1A expression within LUAD, GBMLGG, PAAD, and KIRP TCGA diseases. Fourteen gene sets commonly enriched among the four diseases are shown in the heatmap. B) GSEA was used to identify enriched pathways with genes that are correlated with ERO1α. Heatmap coloring indicates normalized enrichment score (NES). C) GSEA running sum statistic visualizations are shown for the Hallmark gene set, Hypoxia, that was significantly enriched in GBMLGG, KIRP, LUAD, and PAAD TCGA diseases. All common gene sets were Hallmark except for the KEGG “Small Cell Lung Cancer” gene set. GSEAv2.2.3 was used with v6 gene sets sourced from MSigDB. 10,000 gene set permutations were performed using weighted mode scoring and Pearson metric (Subramanian, et al., 2005). Only genes with evidence of expression in > 50 % of a disease patient population were considered.
