Fig. 1.
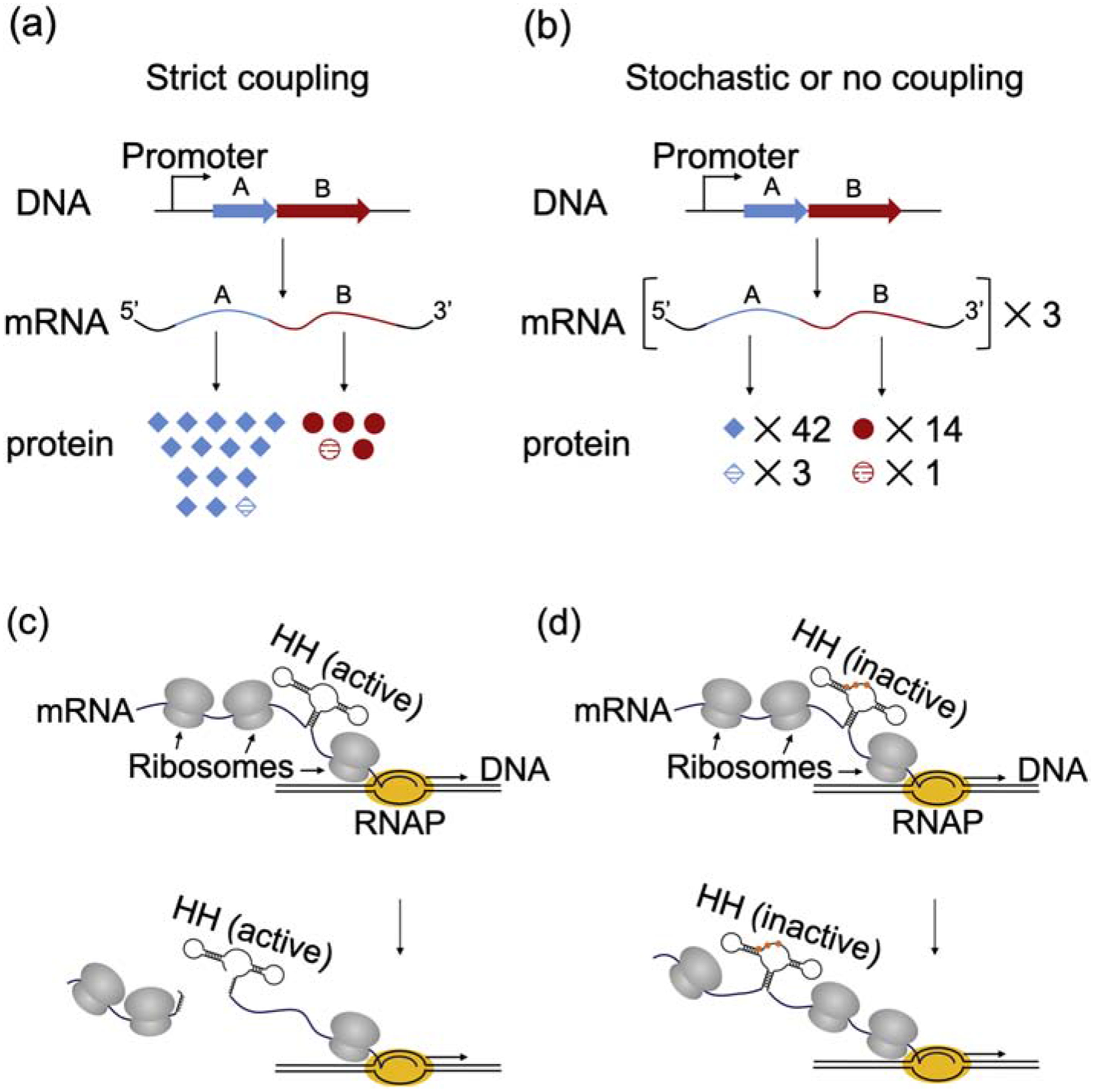
Rationale and approach of Chen and Fredrick (2018). (A-B) Hypothetical protein products of strict versus stochastic coupling. (A) A scenario in which RNAP and the lead ribosome are strictly coupled. Genes A and B are co-transcribed, resulting in stoichiometric levels of A and B mRNA (1:1). The first round of translation by the RNAP-coupled ribosome generates equivalent amounts of proteins A and B (1:1, striped symbols), whereas multiple-round translation yields different amounts of total protein (3:1, all symbols). (B) A scenario in which transcription and translation are uncoupled or stochastically coupled. In this case, RNAP has effectively no impact on the lead ribosome, and hence the ratio of protein products made during transcription (3:1, striped symbols) matches that of multiple-round translation (3:1, all symbols). (C-D) A hammerhead ribozyme enables direct comparison of ‘single’- (co-transcriptional) versus multiple-round translation. (C) The active hammerhead (HH) quickly self-cleaves after being made, so only a ribosome tailgating RNAP will produce full-length LacZ. (D) Three point mutations (colored dots) inactivate the hammerhead without altering the encoded polypeptide, enabling multiple-round translation to be measured.
