Extended Data Fig. 1. Optimization and hits from the liposome leakage assay screen.
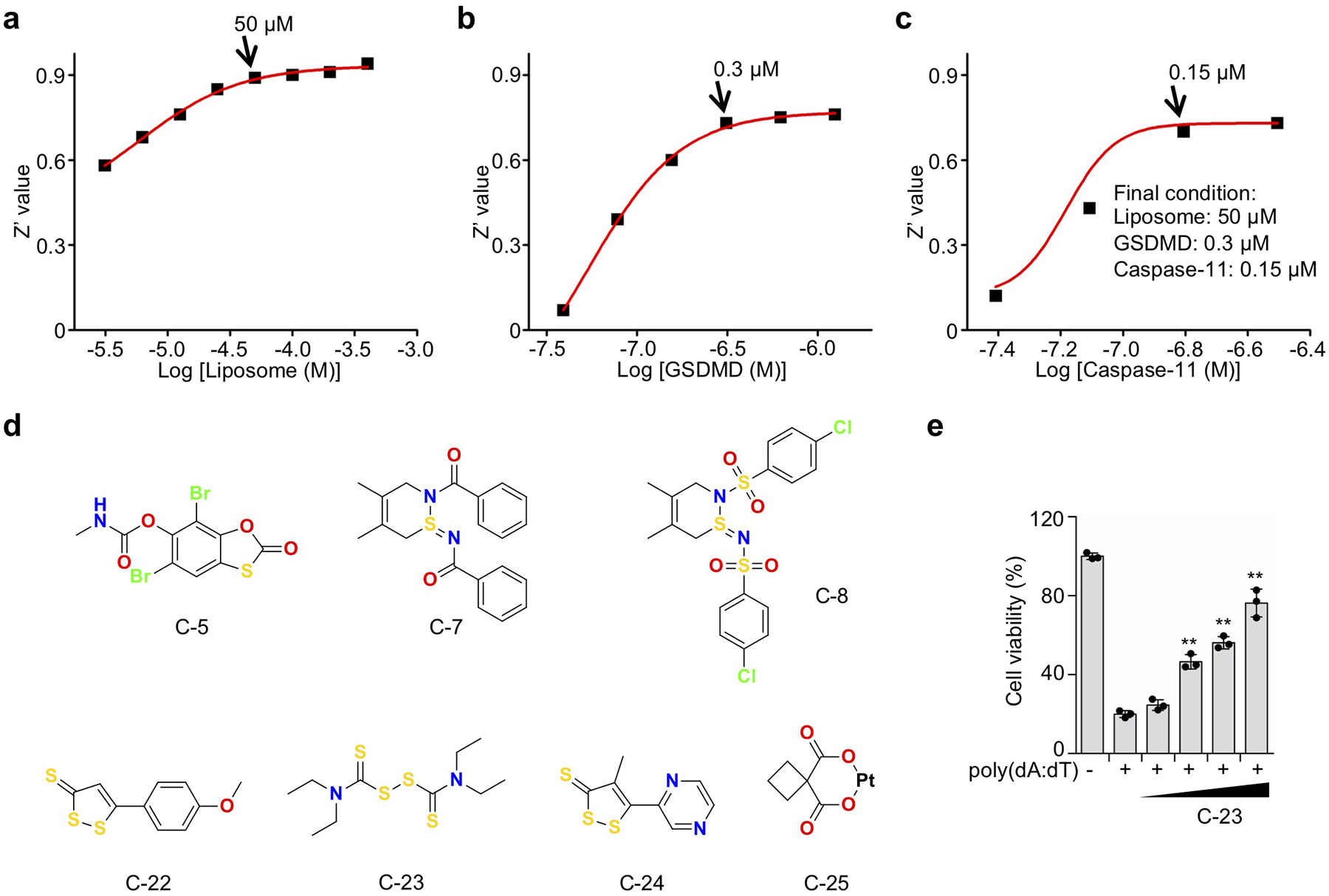
(a-c) Optimization of the Tb3+/DPA assay. (a) GSDMD (2.5 μM) and caspase-11 (2.5 μM) were incubated in liposome solutions at various concentrations in 20 mM HEPES buffer (150 mM NaCl) for 1 hr. The concentration of liposome lipids for the screen was set at 50 μM. n = 3 independent experiments. The mean ± s.e.m. is shown. (b) Different concentrations of GSDMD and caspase-11 (1:1 ratio) were incubated in liposome (50 μM) solutions for 1 hr. The concentration of GSDMD used in the screen was set at 0.3 μM. n = 3 independent experiments. The mean ± s.e.m. is shown. (c) Different concentrations of caspase-11 and GSDMD (0.3 μM) were incubated in liposome (50 μM) solutions for 1 hr. The concentration of caspase-11 used in the screen was set at 0.15 μM. n = 3 independent experiments. The mean ± s.e.m. is shown.The fluorescence intensity at 545 nm was measured after excitation at 276 nm. (d) Hit compounds evaluated in binding and/or cell-based assays. (e) Mouse iBMDMs were pretreated or not with disulfiram (C-23) ranging from 5–40 μM for 1 hr before transfection with PBS or poly(dA:dT) and analyzed for cell viability by CellTiter-Glo assay 4 hrs later. Graphs show mean ± s.d; data are representative of three independent experiments with replicates (n= 3) and similar results. Data were analyzed using two-tailed Student’s t-test. **P < 0.01.
