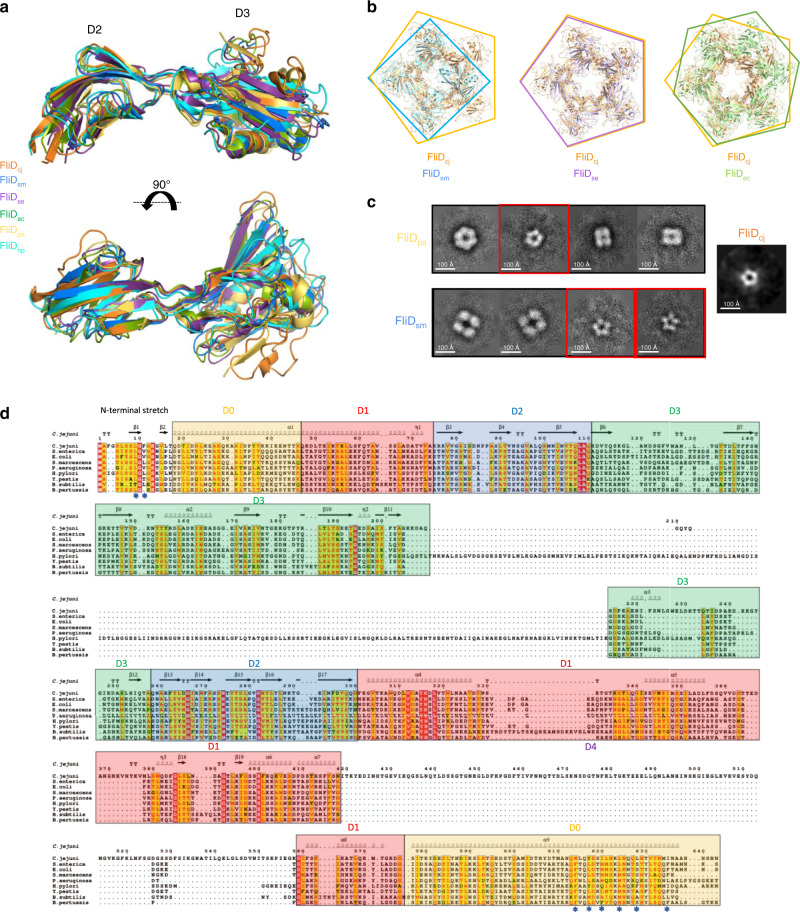
Fig. 2. Comparison of the FliD structures across bacterial species.
a Overlay of the FliD D2–D3 domains structures from C. jejuni (FliDcj, 6SIH—this study, orange), S. marcescens (FliDsm, 5XLJ, blue), S. enterica (FliDse, 5H5T, purple), E. coli (FliDec, 5H5V, green), P. aeruginosa (FliDpa, 5FHY, yellow), and H. pylori (FliDhp, 6IWY, cyan). b Alignments of X-ray crystallography derived oligomeric structures of the head domains from FliDsm, FliDse, and FliDec, to that of the FliDcj pentamer. The diameter of the lumen as well as the outer diameter of the capping protein is similar in the pentamer formed by FliDse, but significantly smaller in the tetramer formed by FliDsm, and larger in the hexamer formed by FliDec. c 2D classes obtained from negative-stain data of recombinant FliDpa and FliDsm (Supplementary Fig. 1b). FliDpa shows three different sets of particles: a class with particles showing 6-fold symmetry (~120 Å in diameter), a class showing smaller particles with 5-fold symmetry (~100 Å diameter), and classes of particles with 4-fold symmetry (~80 Å and ~110 Å in classes 3 and 4, respectively). FliDsm shows two different sets of particles: large particles with 4-fold symmetry (~150 Å in diameter), and smaller particles with 5-fold symmetry (~100 Å diameter). The size of the 5-fold symmetry particles in both orthologues is similar to that observed in 2D classification of FliDcj. The red squares indicate classes with particles of similar shape and dimensions to the FliDcj structure, with distinctive five-fold symmetry. FliDcj negative stain top view is shown as a reference to the right. d Sequence alignment for FliD from different species. The secondary structure elements for FliDcj are shown at the top. The domains are color coded as follows: Yellow: D0 terminal domains. Red: D1 domain. Blue: D2 domain. Green: D3 domain. Uncolored: D4 domain found only in FliDcj. Stars indicate important residues in terminal interactions, for which mutagenesis experiments were performed in Fig. 3.