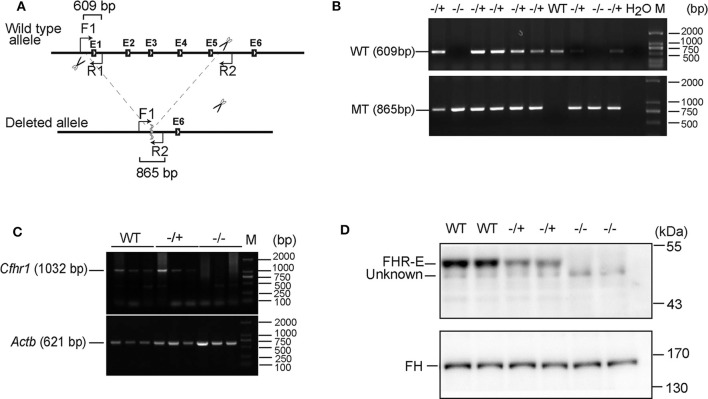
Figure 2.
Verification of the absence of FHR-E in Cfhr1-KO mice. (A) The scheme of Cfhr1 knockout strategy. F1, R1, and R2 indicate the primers used for genotyping. The expected bands amplified by two pairs of primers were 609 and 865 bp, respectively. (B) Verification of Cfhr1 deletion from genome level. The upper bands were amplified with primers F1 and R1. These bands can only be amplified in wild-type and heterozygous mice and cannot be amplified in homozygous mice because of the absence of the sequence of primer R1. The lower bands were amplified with primers F1 and R2. These bands can only be amplified in heterozygous and homozygous mice. The sequence between primers F1 and R2 is too long to be amplified in wild-type mice. (C) Verification of Cfhr1 deletion from RNA level. Hepatic RNA of mice of different genotypes was extracted and reversely transcribed. Primers spanning the open reading frame (ORF, 1,032 bp, NM_015780) of Cfhr1 were used for RT-PCR. Actb was used as a semi-quantitative control. (D) Verification of FHR-E deficiency at the protein level. Plasma proteins of mice with different genotypes were separated by 8% SDS-PAGE gel. Proteins were transferred to the membrane and analyzed by Western blotting using anti-serum to mouse FHR-E, which was generated in rabbit immunized with a recombinant peptide of CCP 3–5 of FHR-E. An about 50 kDa specific FHR-E band was detected in WT and heterozygous mice, but not in homozygous mice. “Unknown” indicates an unknown protein recognized by anti-FHR-E antibody. Western blotting of FH was regarded as an internal control.