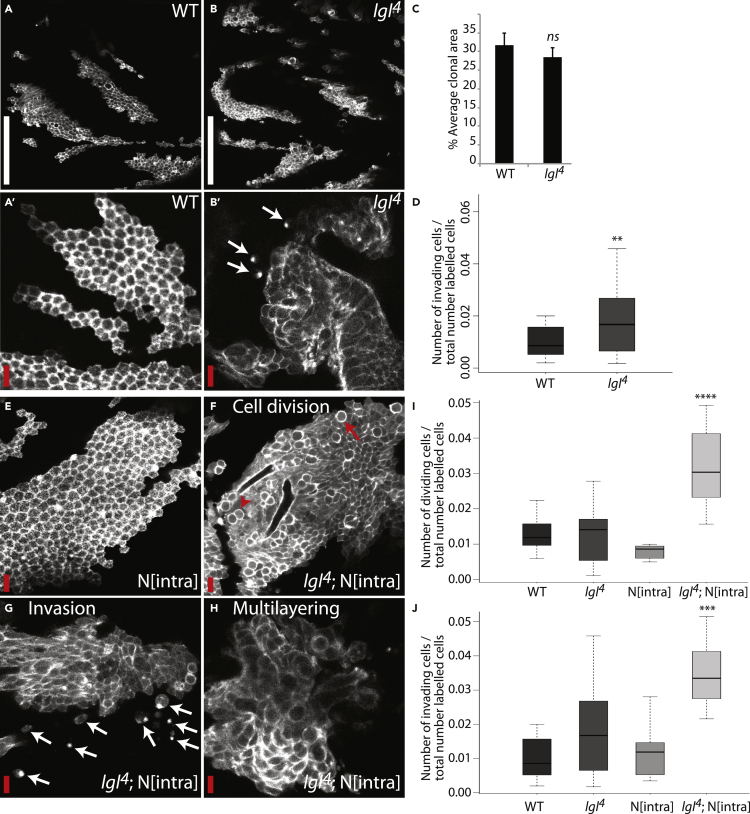
Figure 1.
lgl4 Mutant Clones Provide an Ideal Genetic Background for an Enhancer/Suppressor Screen for Tumor Progression
(A and B) GFP:Moe-labeled genetic clones in the dorsal thorax epithelium of living fly pupae. Clones shown are wild-type (A and A′) or homozygous mutant for the neoplastic tumor suppressor lgl (B and B′).
(C and D) Quantification of average clonal area (C) (n = 10 [WT]; 18 [lgl4]) and the number of invading cells/the total number of labeled cells (D) (n = 30 [WT]; 41 [lgl4]). Quantification shows lgl4 mutant clones to be similar to WT clones in size, with a significant increase in the number of invading cells.
(E–H) GFP:Moe-labeled genetic clones in the dorsal thorax epithelium of living fly pupae. Clones shown are overexpressing activated Notch (Nintra; (E) or simultaneously homozygous mutant for lgl4 and overexpressing Nintra (F–H). Highlighted are effects on cell division (F), invasion (G), and multilayering (H).
(I and J) Quantification of the number of dividing cells (I) and the number of invading cells (J) over the total number of labeled cells for clones with the genotypes shown (n = 30 [WT]; 41 [lgl4]; 7 [Nintra]; 13 [lgl4; Nintra]). Error bars represent ± SEM. Student's t test (E) and Kruskall-Wallis test (F, K, and L) were performed to determine statistical significance. p > 0.05 was considered not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Red arrow, dividing cell; red arrowhead, cell doublet following cytokinesis; white arrows, invading cells. White scale bar, 50 μm; red scale bar, 10 μm.