Figure 6.
SA1 or SA2KD Promotes Invasion
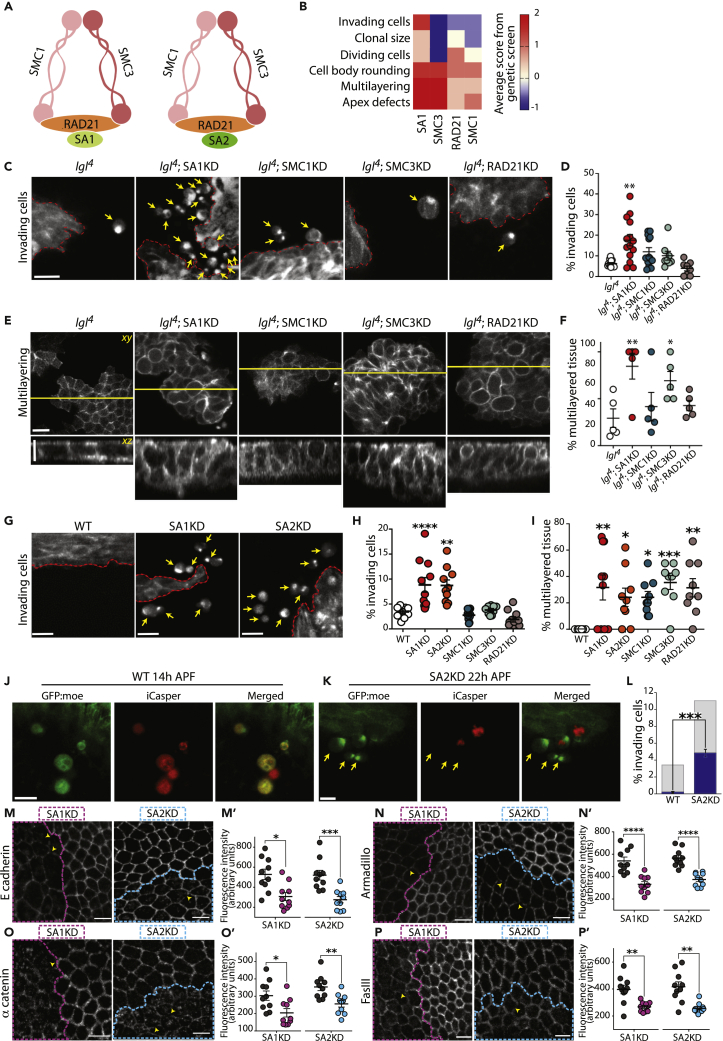
(A) Somatic cells simultaneously express two different Cohesin rings, differentiated by the presence of either SA1/STAG1 or SA2/STAG2.
(B) Heatmap illustrating qualitative scores given to cohesin subunits included in the genetic screen. A subset of categories is shown. Red, enhancement of a phenotype; yellow, no phenotype change; blue, inhibition of a phenotype.
(C–F) GFP:moe positively marked lgl4 mutant clones with additional cohesin complex subunit KD, showing invading cells (arrows; (C)) and multilayering (E), quantified in (D) and (F); n = 5 animals/genotype. Red dashed line highlights edge of clone. Yellow line shows position of xz slice shown.
(G) Basal confocal slice of GFP:moe positively marked WT, SA1, or SA2KD clones, highlighting invading cells (arrows).
(H and I) Quantification of % invading cells (H) and % multilayering (I) following KD of each cohesin subunit, compared with WT.
(J–L) Confocal images of the basal surface of iCasper (red) and GFP:Moe (green)-labeled WT clones (J) and SA2KD clones (K). Arrows highlight invading cells that are iCasper negative. Quantified in (L): Gray, % invading cells/total number of labeled cells; blue, % non-apoptotic invading cells/total number of labeled cells; n = 50 cells from 10 animals/genotype. Young WT pupae were used as a control (J) as older WT animals have little to no invading cells.
(M–P) SA1 or SA2KD clones, highlighted by magenta and cyan dashed lines, respectively, show disrupted E-cadherin (M, quantified in M'), armadillo (N, quantified in N'), α-catenin (O, quantified in O'), and fasIII (P, quantified in P'), localization. Arrowheads highlight junctional breaks. Quantification shows fluorescence intensity at the level of the junction (n = 100 junctions from 10 animals for each genotype). Scale bars, 10 μm. Error bars = ± SEM. Student's t test or one-way ANOVA with Dunnett's post hoc test for multiple comparisons was performed to determine statistical significance. p > 0.05 was considered not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figures S5 and S6.