Figure 5. Validation of the hippocampal MGE translatome between Ribohet and Ribohomo alleles using iDEP9.0.
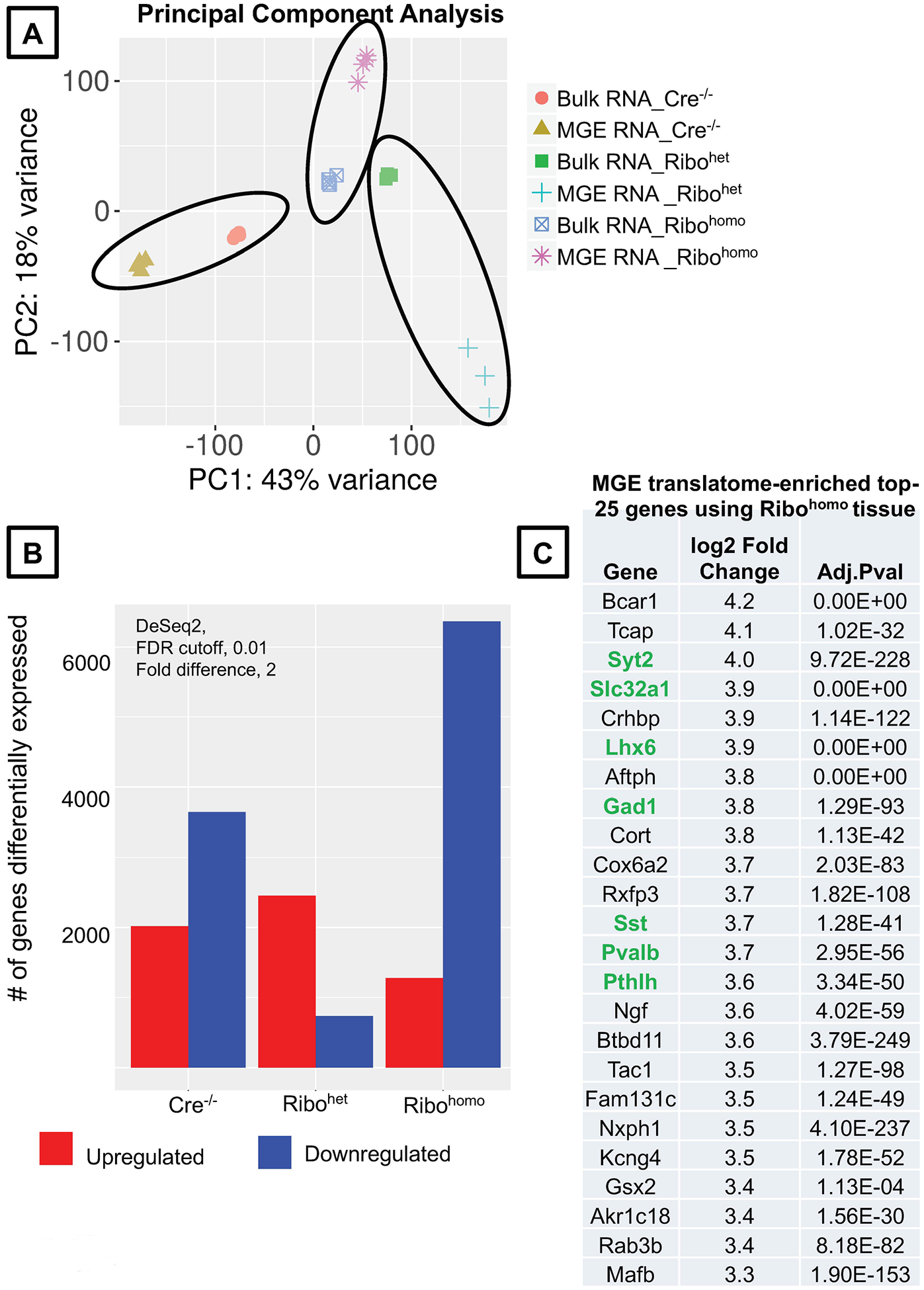
(A) Principal component analysis plot comparing the Bulk RNA vs MGE-specific Ribotag-associated RNA across different genotypes. (B) Differentially up and downregulated genes according to DeSEq2 at a stringent FDR <0.01 and fold difference >2, in the MGE-specific Ribotag-associated translatome. (C) MGE-translatome-enriched, top 25 genes obtained from Rpl22HA/HA RNAsequencing. Genes highlighted in green are established MGE-enriched positive controls (see Supplemental Table. 1 for the complete list of differentially enriched genes identified in the present Ribohomo Ribotag assay).
