Abstract
The joining of interruptions in the phosphodiester backbone of DNA is critical to maintain genome stability. These breaks, which are generated as part of normal DNA transactions, such as DNA replication, V(D)J recombination and meiotic recombination as well as directly by DNA damage or due to DNA damage removal, are ultimately sealed by one of three human DNA ligases. DNA ligases I, III and IV each function in the nucleus whereas DNA ligase III is the sole enzyme in mitochondria. While the identification of specific protein partners and the phenotypes caused either by genetic or chemical inactivation have provided insights into the cellular functions of the DNA ligases and evidence for significant functional overlap in nuclear DNA replication and repair, different results have been obtained with mouse and human cells, indicating species-specific differences in the relative contributions of the DNA ligases. Inherited mutations in the human LIG1 and LIG4 genes that result in the generation of polypeptides with partial activity have been identified as the causative factors in rare DNA ligase deficiency syndromes that share a common clinical symptom, immunodeficiency. In the case of DNA ligase IV, the immunodeficiency is due to a defect in V(D)J recombination whereas the cause of the immunodeficiency due to DNA ligase I deficiency is not known. Overexpression of each of the DNA ligases has been observed in cancers. For DNA ligase I, this reflects increased proliferation. Elevated levels of DNA ligase III indicate an increased dependence on an alternative non-homologous end-joining pathway for the repair of DNA double-strand breaks whereas elevated level of DNA ligase IV confer radioresistance due to increased repair of DNA double-strand breaks by the major non-homologous end-joining pathway. Efforts to determine the potential of DNA ligase inhibitors as cancer therapeutics are on-going in preclinical cancer models.
Introduction
Interruptions in the phosphodiester backbone pose serious threats to genome integrity and cell viability. While these can be generated directly by DNA damaging agents such as ionising radiation, they are also produced as intermediates during normal DNA metabolism. This includes single-strand breaks that occur between Okazaki fragments during DNA replication and site-specific DNA double-strand breaks that are generated during meiosis and V(D)J recombination. In addition, DNA single-strand breaks are generated during the correction of replication errors, excision of DNA damage, somatic hypermutation and removal of DNA methylation during cell differentiation. These breaks are all ultimately sealed by a DNA ligase encoded by one of the three human LIG genes (Figure 1).
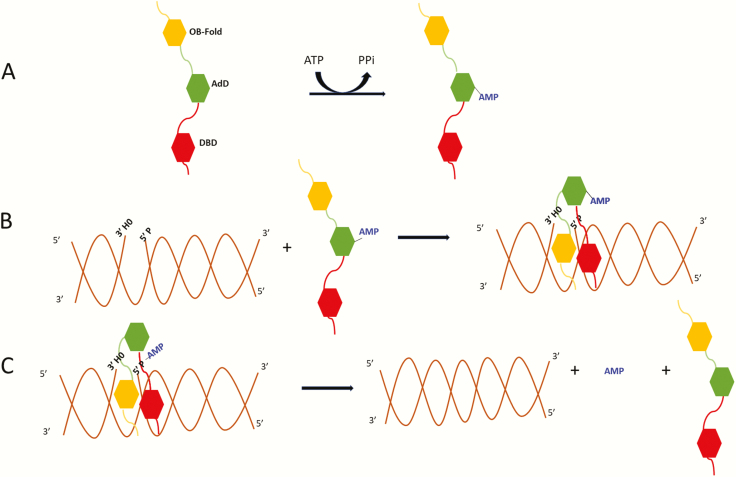
Fig. 1.
Three-step ligation reaction. (A) DNA ligases in an extended conformation interact with ATP to generate a covalent AMP-ligase intermediate with the AMP moiety linked to specific lysine residues in the Adenylation Domain (AdD, green). (B) When the DNA ligase recognises a ligatable nick, the catalytic region composed of the DNA Binding Domain (DBD, red) and Oligonucleotide/Oligosaccharide Binding-fold Domain (OBD, yellow) in addition to the AdD changes conformation, encircling the DNA nick with each of the three domains contacting the DNA. Within this structure, the covalently bound AMP is transferred to the 5′ termini of the nicked DNA. (C) Lastly, using the OH group at the 3′ termini as a nucleophile, non-adenylated DNA ligase catalyses the phosphodiester bond formation releasing the bound AMP.
The DNA ligases encoded by the human LIG1, LIG3 and LIG4 genes belong to the nucleotidyl transferase family that also includes RNA ligases and mRNA capping enzymes (1,2). In the first step of the ligation reaction (Figure 1), ATP is used as the co-factor by the human DNA ligases to generate a covalent ligase-AMP intermediate in which the AMP moiety is covalently linked to a lysine residue (3), although a recent report provided evidence that DNA ligase IV can also utilise NAD as the adenylation donor (4). Subsequently, the AMP moiety is transferred from the active site lysine of the ligase polypeptide to the 5′ phosphate terminus at a DNA break. In the final step, the non-adenylated ligase engages with DNA adenylate and, utilising the hydroxyl at the 3′ terminus as a nucleophile, catalyses phosphodiester bond formation and release of the AMP moiety (3). While the three steps of the ligation reaction are normally coordinated, it is now evident that under some circumstances the potentially cytotoxic DNA adenylate intermediate is released, contributing to neuronal cell death in the inherited human neurodegenerative disease, ataxia oculomotor apraxia-1 (5).
The LIG1, LIG3 and LIG4 genes each encode a DNA ligase that functions in nuclear DNA metabolism (3). In contrast, mitochondria contain a single species of DNA ligase that is encoded by the LIG3 gene and is essential for mitochondrial DNA replication and repair (6–9). Insights into the functions of the three nuclear DNA ligases have been gleaned from a combination of genetic and biochemical approaches, in particular the identification of partner proteins that predominantly interact with the regions flanking the conserved catalytic region of the DNA ligases (Figure 2). Both nuclear DNA ligase IIIα and DNA ligase IV exist in stable complexes with partner proteins, XRCC1 and XRCC4 (Figure 2), respectively, that are required for the stability and activity of the DNA ligase as well as providing additional binding interfaces for other protein–protein interactions (10–13). While a number of interacting proteins have been identified for DNA ligase I, none of these partners appear to influence the stability of DNA ligase I (14–16).
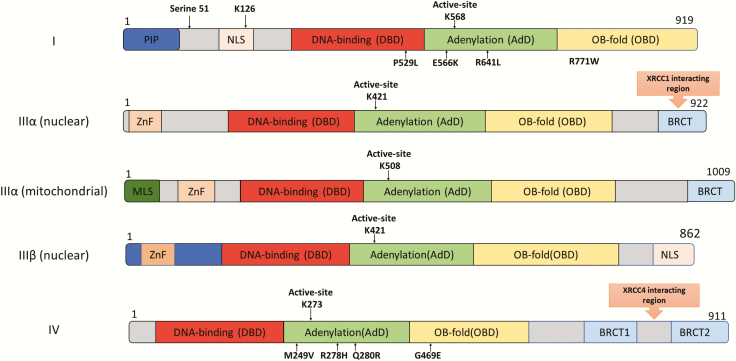
Fig. 2.
Domain organisation of the DNA ligases encoded by the human LIG1, LIG3 and LIG4 genes. The Adenylation Domain (AdD, green) and Oligonucleotide/Oligosaccharide Binding-fold Domain (OBD, yellow) domains comprise the catalytic core that contains the key active site lysine residue. The less conserved DNA Binding Domain (DBD, red) is N-terminal to this core. The non-catalytic N-terminal region of DNA ligase I contains PCNA interaction peptide, PIP (blue) and nuclear localisation signal (beige). The three isoforms of DNA ligase III have an N-terminal Zinc finger domain (light orange: ZnF). Mitochondrial DNA ligase IIIα has a mitochondrial localisation signal, MLS (dark green) at the N-terminus. DNA ligase IIIα and DNA ligase IV contain one and two C-terminal BRCT domains (blue), respectively. The DNA ligase IIIα BRCT domain is required for interaction with XRCC1 and whereas XRCC4 interacts with the region between the DNA ligase IV BRCT domains. Amino acid substitutions identified in DNA ligase deficiency syndromes are indicated below the DNA ligase polypeptides.
It is now evident that there is significant functional overlap between the three DNA ligases in nuclear DNA transactions. For example, while there is substantial evidence indicating that DNA ligase I is the predominant enzyme joining Okazaki fragments during DNA replication (3), the LIG1 gene is not essential in either chicken DT40 or mouse cells (17–20). However, DNA ligase I appears to be essential in rapidly proliferating human cancer cells and also has unique functions in post-replicative repair, including the generation of sister chromatid telomere fusions (21). In the yeast, Saccharomyces cerevisiae, the LIG1 homolog CDC9 is essential but a notable difference between S. cerevisiae and vertebrates is that S. cerevisiae lacks a homolog of the LIG3 gene (22). At least in mouse and chicken DT40 cells, it is evident that DNA ligase IIIα joins Okazaki fragments in the absence of DNA ligase I (20,23). This redundancy also occurs in base and nucleotide excision repair and probably single-strand break repair with DNA ligase IIIα and DNA ligase I participating in different sub-pathways of repair synthesis and ligation (3,24–27). DNA ligase IV-dependent non-homologous end joining is the major pathway for repairing DNA double-strand breaks (DSB)s, particularly in the G1 phase of the cell cycle and in non-dividing cells (28). Recombinational repair makes a significant contribution to DSB repair in the S and G2 phases of the cell cycle when sister chromatids are available as templates to guide the repair. It is likely that DNA ligase IIIα and DNA ligase I are both involved in the completion of recombinational repair. These enzymes also participate in alternative non-homologous end-joining (a-NHEJ) pathways that serve as back-ups for the major DNA ligase IV-dependent NHEJ pathway and recombinational repair (29–31). It is likely that the extent of functional redundancy between the DNA ligases in DNA replication and repair varies between cell types and species, giving rise to apparently contradictory conclusions regarding the cellular functions of the DNA ligases.
In this review, we summarise the links between DNA ligases and human disease. Inherited mutations in LIG1 and LIG4 have been identified as the causative factors in human immunodeficiency syndromes whereas altered DNA ligase expression and/or activity have been observed in cancer, serving as biomarkers of altered repair and resistance to therapy, and neurodegeneration.
Human DNA ligase I
DNA ligase I polypeptide
The gene encoding DNA ligase I was the first of three human LIG genes to be identified in 1990. Human cDNAs that complemented the temperature sensitive growth of a S. cerevisiae cdc9 mutant strain were identified and confirmed to encode DNA ligase I by the presence of regions homologous to the sequence of tryptic peptides from purified bovine DNA ligase I (32). DNA ligase I is a 919 amino acid polypeptide (Figure 2) with a molecular weight of 102,000 although it has an apparent molecular mass of 125 kDa determined by SDS-polyacrylamide gel electrophoresis because of a high proline content that causes anomalous mobility (32). The C-terminal region (residues 536–919) contains six conserved motifs within Adenylation (AdD; residues 536–748, Figure 2) and the Oligonucleotide/Oligosaccharide Binding-fold Domains (OBD; residues 749–919, Figure 2) that are also present in other members of the nucleotidyl transferase family (2,32,33). The conserved motif that contains the key active site lysine residue of the nucleotidyl transferase family was identified by the sequencing of the adenylated tryptic peptide from bovine DNA ligase I (34). In human DNA ligase I, the active site lysine is Lys568 (Figure 2). Crystallisation of an active fragment of human DNA ligase I (residues 233–919) with a non-ligatable nicked DNA substrate led to the identification of an additional domain N-terminal to the AdD that is less well conserved amongst eukaryotic DNA ligases compared with the AdD and OBD (35,36). This domain, which binds more robustly to DNA than the AdD and OBD, is referred to as the DNA binding domain (DBD, Figure 2). In the atomic resolution structure (Figure 3a), the DBD, AdD and OBD encircle the nicked DNA with each domain contacting the DNA duplex and the AMP moiety attached to the 5′ termini of the nick with the AdD (35). While the N terminal region of DNA ligase I is not involved in catalysis, it contains the nuclear localisation signal, is post-translationally modified and participates in protein–protein interactions (37–42). At the very N-terminus, there is a PCNA Interacting Peptide motif or PIP box that directly binds to PCNA, an interaction that is critical for recruitment to replication foci (14,40,43). In addition, the DNA ligase I N-terminal region is involved in a phosphorylation-regulated interaction with replication factor C (44), which loads the PCNA ring onto DNA and a methylation-dependent interaction with UHRF1 (45), a key component of the maintenance DNA methylation machinery.
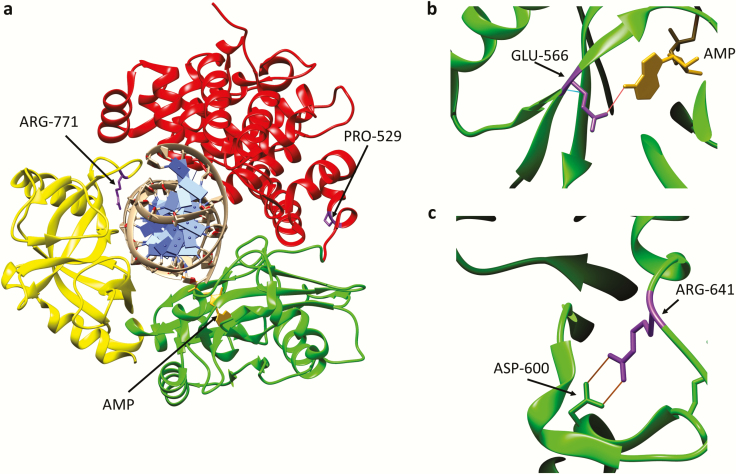
Fig. 3.
Amino acid substitutions identified in individuals with DNA ligase I deficiency syndrome. (a) Ribbon diagram showing the Adenylation domain (AdD, green), OB-fold domain (OBD, yellow) and DNA binding domain (DBD, red) of DNA ligase I encircling a nicked DNA duplex (grey). The AMP group (gold) linked to the 5-phosphate terminus of the DNA nick held within the AdD is indicated. Substitution of Arg771 (purple) in the OBD alters interaction with DNA, resulting in reduced enzymatic activity. with Trp and Pro529 depicted in purple. Substitution of Pro529 (purple) in the DBD with Leu has no effect on enzymatic activity. (b) Glu566 residue (purple) forms a hydrogen bond with N6 of the AMP moiety (gold). Replacement of Glu566 Lys inactivates enzyme activity. (c) Arg641 (purple) forms a salt bridge with Asp600 within the AdD domain. Replacement of R641 with Leu disrupts the salt bridge, resulting in reduced enzymatic activity that appears to be due to defective DNA binding.
DNA ligase I and human disease
The first known case of inherited DNA ligase I deficiency in humans was described in 1992. This female individual was underweight and anaemic at birth and continued to exhibit growth retardation and developmental delays as well as sensitivity to sunlight. In addition, the individual had recurrent ear and chest infections, indicating immunodeficiency, and died at age 19 from pneumonia (46,47). More recently, another five individuals with inherited DNA ligase I deficiency have been identified. Novel LIG1 mutations as well as one of the mutations identified in the first case were found in these individuals who exhibited a range of clinical symptoms, including immunodeficiency (48). The first individual inherited a mutant allele resulting in the substitution of Arg771 within the DBD with Trp and had a second mutant allele of unknown origin resulting in the replacement of Glu566 with Lys (46) (Figure 2). Notably, in the second group, three of the individuals were homozygous for a mutant allele in which Arg771 was replaced with Trp and Pro529 was replaced with Leu (Figures 2 and 3a). The other two individuals inherited two mutant alleles, one of which results in a truncated polypeptide terminating at Thr415 with the other encoding a full length polypeptide in which Arg641 is replaced with Leu (48). The truncated polypeptide is inactive as it lacks the catalytic region. Similarly, the E566K polypeptide is also inactive as Glu566 forms a key hydrogen bond with the N6 of adenine in the ATP co-factor and so, the mutant polypeptide is unable to form the ligase-AMP intermediate (35,48) (Figure 3b). While the replacement of Pro529 with Leu does not impact catalytic activity, both the R771W and the R641L polypeptides have significantly reduced activity, resulting in accumulation of the DNA-adenylate intermediate (46,48,49). Arg771 within the DBD directly interacts with the template strand of the nicked DNA (Figure 3a) and so is important for DNA binding, whereas Arg641 in the AdD is involved in a salt bridge linking two alpha helices (Figure 3c) that presumably stabilises a conformation that binds to DNA (35).
A primary fibroblast cell line, 46BR, and a SV-40 transformed subline (46BR.1G1) were established from the first individual with inherited DNA ligase-I deficiency with 46BR.1G1 only expressing the R771W version of DNA ligase I (46). Compared to SV40-transformed-fibroblasts with wild-type DNA ligase I, 46BR.1G1 cells showed increased sensitivity to a range of DNA damaging agents, in particular DNA alkylating agents (50,51). In addition, they also had an increased incidence of sister chromatid exchange, delayed rejoining of strand breaks, elevated levels of phosphorylated H2AX and defects in Okazaki fragment ligation but were reported to be hypomutable by DNA damaging agents (50–54). Similarly, lymphoblastoid B cells and peripheral blood T cells expressing the R641L version isolated from DNA ligase I-deficient individuals also exhibited increased sensitivity to DNA alkylating agents and elevated levels of phosphorylated H2AX following ionising radiation (48).
A mouse model expressing a version of murine DNA ligase I equivalent to human R771W DNA ligase I exhibited delayed growth, reduced erythropoiesis leading to enlargement of the spleen, increased genomic instability and increased incidence of spontaneous epithelial tumours (55). In contrast to human DNA ligase I-deficient cells, embryonic fibroblasts established from the mouse model did not exhibit increased DNA damage sensitivity (50,51,55), suggesting that the increased genomic instability and cancer predisposition in the mouse may be due to genomic rearrangements arising from DNA replication defects. At the present time, there is no convincing evidence that DNA ligase I deficiency in humans results in increased cancer predisposition but it is clear that it causes immunodeficiency (46,48). Interestingly, DNA ligase I-deficient individuals have a wide range of abnormalities in blood cells including hypogammaglobulinemia, increased levels of circulating immature T cells, lymphopenia and erythrocyte macrocytosis, suggesting that defects in replicative DNA synthesis may selectively impact rapidly dividing cell populations, such B and T cells, in humans (46,48).
Steady-state levels of DNA ligase I protein were significantly higher in 29 different malignant tumour samples compared to the benign tissue samples obtained from human patients (56), suggesting that overexpression of DNA ligase I is a common feature of cancer cells. This is not surprising, since studies in cell culture models have shown that expression of the LIG1 gene correlates with proliferation and cancer cells tend to be highly proliferative (56,57). Thus, the expression levels of DNA ligase I are likely to be indicative of the proliferative status of the cancer cell. Since knockdown of DNA ligase I with antisense oligonucleotides inhibited the growth of MCF-7 cancer cells (56) and DNA ligase I appears to have essential functions in a human cancer cell line (21), DNA ligase I may be a good target for the development of cancer therapeutics. It is, however, likely that DNA ligase I inhibitors will impact immune function and highly proliferating normal tissues and cells in addition to cancer cells.
Human DNA ligase III
DNA ligase III polypeptides
Two groups using different approaches reported the identification of the LIG3 gene in 1995 (58,59). While these groups described mRNAs with different 3′ ends, they both showed that an internal ATG within the open reading frame was the preferred translation initiation site resulting in the synthesis of 862 and 922 amino acid polypeptides referred to as DNA ligase IIIβ and DNA ligase IIIα, respectively (Figure 2). A subsequent study revealed that the shorter mRNA encoding DNA ligase IIIβ is generated by an alternative splicing event that thus far has only been detected in male germ cells (60). The Campbell laboratory noticed that the open reading frame following the first ATG in DNA ligase IIIα mRNA encodes an amphipathic helix that serves as a mitochondrial targeting sequence and showed that mitochondrial and nuclear versions of DNA ligase IIIα are generated by alternative translation initiation in somatic cells (6,61). Although mitochondrial DNA ligase IIIα is larger than the nuclear form (1009 amino acids versus 922 amino acids), these polypeptides are ultimately very similar in size following the proteolytic removal of the mitochondrial targeting sequence by the mitochondrial protein import machinery. In the absence of a nuclear localisation signal, it appears likely that nuclear DNA ligase IIIα forms a complex with XRCC1 in the cytoplasm and utilises the nuclear localisation signal of XRCC1 to enter the nucleus. This interaction, which occurs between the BRCT domains at their C-termini (Figure 2), results in the formation of a stable nuclear DNA ligase IIIα/XRCC1 heterodimer (12,13,60,62). Since XRCC1 interacts with several other DNA repair enzymes, it has been suggested that it serves as scaffold for the assembly of functional multiprotein DNA repair complexes containing DNA ligase IIIα (13). Interactions of DNA ligase IIIα and, in particular, XRCC1 with poly (ADP-ribosylated) PARP1 are critical for the recruitment of DNA ligase IIIα/XRCC1 and XRCC1-interacting proteins to single-strand breaks (26,63). TDP1, which is involved in the removal of trapped topoisomerase 1 complexes, is the only protein that interacts with nuclear DNA ligase IIIα but not XRCC1 identified so far (64). In the mitochondria, DNA ligase IIIα functions in the replication and repair of mitochondrial DNA independently from XRCC1, which is absent from this organelle (61).
In contrast to DNA ligase I, DNA ligase III polypeptides contain robust DNA binding activities (65). An N-terminal zinc finger, which is similar to the two N-terminal zinc fingers of PARP1, forms a DNA binding module with the DBD that binds to strand breaks irrespective of the structure of the termini (65–67). In addition, the AdD and OBD also act together to bind to ligatable DNA nicks (65,67). Based on these observations, it has been proposed that the N-terminal zinc finger-DBD module act as an initial strand break sensor that is displaced by the AdD and OBD if the break has ligatable termini (65,67). In addition, the zinc finger enhances the intermolecular joining of duplex DNA ends (68). The atomic resolution structure of the catalytic region of DNA ligase III (DBD, AdD and OBD) determined in complex with non-ligatable nicked DNA is very similar to that determined for DNA ligase I, indicating that, despite the differences in DNA binding, the mechanism of nick engagement immediately prior to ligation is conserved (35,67).
DNA ligase III and human disease
Unlike LIG1 and LIG4, no inherited human syndrome has been linked with the LIG3 gene. Interestingly, mutations in the XRCC1 gene and in genes encoding the XRCC1-interacting proteins aprataxin and PNKP, and the DNA ligase IIIα-interacting protein, TDP1, have been identified as the causative factor in inherited neurodegenerative syndromes (69). This suggests that DNA ligase IIIα-dependent repair of DNA breaks in the nucleus is critical for neuronal cell viability. The absence of a syndrome associated with LIG3 mutations is likely due to the essential role of DNA ligase IIIα in mitochondria (8,9). By targeting a heterologous DNA ligase to mitochondria, it has been possible to generate cell lines that lack nuclear DNA ligase IIIα. With the possible exception of UV light, these lines do not exhibit significant sensitivity to DNA damaging agents, likely due to functional redundancy with DNA ligase I (8,9). Notably, DNA ligase IIIα catalyses the limiting step in mitochondrial base excision repair and mitochondrial extracts prepared from the brains of Alzheimer's patients have lower levels of mitochondrial DNA ligase IIIα, suggesting that a deficiency in DNA ligase IIIα may underlie the abnormal mitochondrial function observed in Alzheimer's disease and related dementias (70,71).
In cancer, elevated steady-state levels of DNA ligase IIIα as well as PARP1 have been identified as biomarkers of altered DNA double-strand break repair, in which DNA double-strand breaks are channelled away from the major DNA ligase IV-dependent NHEJ and into the a-NHEJ pathway in breast cancer, leukaemias and neuroblastoma, both in cell lines and patient samples (72–74). The changes in gene expression appear to be driven by the c-Myc oncogene (75). Intriguingly, the alteration in DNA double-strand break repair is exacerbated in therapy-resistant disease, suggesting that the change in DNA repair pathway utilisation is part of the response of the cancer cell to acquire resistance to chemotherapy (72,73). The alteration in DNA repair does, however, constitute an opportunity to develop cancer cell-specific therapeutic strategies as cells with the DNA repair alteration are hypersensitive to a combination of PARP and DNA ligase III inhibitors (72,73). While the inhibitor combination does inhibit the repair of DNA double-strand breaks by a-NHEJ, the effect of the DNA ligase IIIα inhibitor appears to be predominantly mediated via inhibition of mitochondrial DNA ligase IIIα in cancer cell mitochondria, suggesting that the inhibitor combination targets both nuclear and mitochondrial DNA metabolism (76).
Human DNA ligase IV
DNA ligase IV polypeptide
The gene encoding DNA ligase IV was identified at the same time as the gene encoding DNA ligase III in a screen for cDNAs encoding sequences homologous to the most C-terminal of the conserved motifs in the nucleotidyl transferase family (59). A unique feature of DNA ligase IV is the tandem array of two C-terminal BRCT domains (Figure 2). The first insights into the function of DNA ligase IV came from an elegant study by the Lieber laboratory showing that DNA ligase IV stably associated with XRCC4, a protein known to be involved in the repair of DNA double-strand breaks (DSB)s by NHEJ and V(D)J recombination in B cells (10,28,77). XRCC4 interacts with the region between the BRCT motifs (Figure 2) and is required for the stability and activity of DNA ligase IV (10,77,78). The repair of DSBs is initiated by the binding of ring-shaped Ku heterodimer to the DNA end with the Ku-DNA complex serving as a platform for the recruitment of other NHEJ proteins including DNA PKcs and DNA ligase IV/XRCC4. While these three core components are sufficient to join duplex DNA ends with cohesive ligatable termini in vitro, PAXX and XLF, which are both XRCC4 homologs, stimulate ligation by DNA ligase IV/XRCC4, presumably via interactions with XRCC4 (79). It has been suggested that XLF and PAXX, which are functionally redundant, act to stabilise juxtaposed DNA ends with damaged and/or incompatible termini (79). These ends require processing by DNA polymerases, such as the Pol X family polymerases µ and λ and nucleases, such as Artemis, prior to ligation (79). During V(D)J recombination, the core components, Ku, DNA PKcs and DNA ligase IV/XRCC4, are required for the generation of coding joints from RAG-initiated DSBs with contributions from some, if not all, of the other NHEJ proteins (79). During NHEJ and V(D)J recombination, the DNA ends are juxtaposed by protein–protein interactions between DNA PKcs molecules on the two ends (80). Given the size of DNA PKcs and its position when assembled with Ku on a DNA end, it appears likely that DNA PKcs must be displaced to allow processing and joining of the ends. The participation of DNA ligase IV/XRCC4 during the initial assembly of the NHEJ proteins at a DNA end and its required role in the last step, suggests that DNA ligase IV/XRCC4 may play a role in the transition between different NHEJ complexes during the repair of DSBs (81–83).
Compared with the other DNA ligases, the re-adenylation of DNA ligase IV molecules after one cycle of catalysis is very slow (84,85). The ability of XLF to modestly increase the turnover of DNA ligase IV in vitro (85), suggests that XLF and possibly other NHEJ factors, such as PAXX, may have a specific role in enhancing re-adenylation of DNA ligase IV. In addition, a recent intriguing study reported that DNA ligase IV is capable of utilising NAD and degradation products of poly (ADP-ribose) as AMP donors in multiple turnover reactions with these novel co-factors binding to the BRCT1 domain of DNA ligase IV (4). Since the joining of DSBs requires two ligation events, a key question is whether one or two DNA ligase IV/XRCC4 complexes are involved. The recent structure of DNA ligase IV complexed with nicked DNA (86) indicates that the catalytic mechanism is similar to DNA ligases I and III (35,67) and so ligation of two closely opposed nicks on opposite strands by one DNA ligase IV/XRCC4 complex would likely require release of the complex after one ligation and re-engagement in the opposite orientation. Alternatively, in a model supported by small angle X-ray scattering analysis of NHEJ complexes, there may be two DNA ligase IV/XRCC4 complexes, one on each end, with their flexible catalytic regions able to engage the nicks on opposite strands (83).
DNA ligase IV and human disease
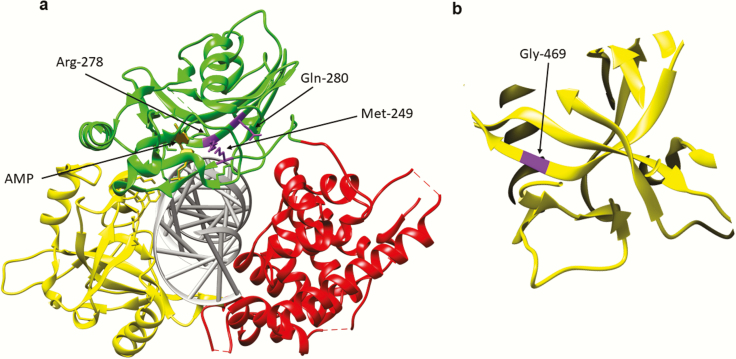
Deletion of the murine LIG4 gene causes embryonic lethality in mice that appears to be due to neuronal cell death via a p53-dependent apoptotic pathway (87–89). While genetic inactivation of p53 results in viable LIG4 null embryos, the resulting mice have defects in growth and lymphocyte development (87). As expected based on the embryonic lethality observed in mice, humans lacking DNA ligase IV activity have not been identified. There have, however, been a larger number of individuals identified with inherited mutant alleles of LIG4 that encode polypeptides with partial activity compared with the other LIG genes. Although these individuals are all considered to have DNA ligase IV syndrome, there is significant variation in the degree of the growth defects, microcephaly, radiosensitivity, chromosomal instability, immunodeficiency and predisposition to malignancy exhibited by these individuals (90) that presumably reflects differences in the DNA ligase IV defect conferred by the inherited mutation. For example, the first identified case of DNA ligase IV syndrome was an individual with a homozygous mutation resulting in replacement of Arg278 with His (Figure 4a) that leads to radiosensitivity but not immunodeficiency (91). Thus, while this amino acid change resulted in a defect in formation of the enzyme-adenylate, it appears that it has sufficient residual activity for V(D)J recombination in lymphocytes, but not enough to repair the larger number of DSBs caused by ionising radiation (91). Additional amino acid changes that cluster near the ATP binding site have been identified in other individuals with DNA ligase IV deficiency syndrome (Figure 4a). Two siblings with severe combined immunodeficiency and microcephaly inherited two compound heterozygous mutations (Q280R and a frameshift at Lys424). The amino acid change at Gln280 is near the conserved active site and likely affects ATP binding pocket (Figure 4a), whereas truncation at Lys424 terminates the polypeptide before the XRCC4 interacting domain required for stable complex formation (92). Another individual presented with radiosensitivity, immunodeficiency and microcephaly along with Epstein bar virus-associated large B-cell lymphoma. Compound heterozygous alleles encoding an amino acid substitution M249V near the ATP binding site (Figure 4a) as well as a frameshift at Lys424 were identified (93). Additional LIG4 mutations that result in truncated polypeptides that are likely to have altered binding with XRCC4 and a mutant version G469E (Figure 4b), that may have a DNA binding defect, have been found in individuals with abnormal facial features, developmental and/or growth delays, microcephaly, pancytopenia and skin aberrations (94). Because of the range and diversity of symptoms caused by mutations in the LIG4 gene, it is difficult to distinguish individuals with DNA ligase IV syndrome from individuals with other chromosomal instability syndromes such as Nijmegen Breakage Syndrome, Seckel syndrome and Fanconi anaemia (94,95).
Fig. 4.
Amino acid substitutions identified in individuals with DNA ligase IV deficiency syndrome. (a) Ribbon diagram showing the Adenylation domain (AdD, green), OB-fold domain (OBD, yellow) and DNA binding domain (DBD, red) of DNA ligase IV encircling a nicked DNA duplex (grey). The AMP group (gold) linked to the 5-phosphate terminus of the DNA nick held within the AdD is indicated. Amino acid substitutions identified in the AdD; R278H, Q280R, M249V (purple) are indicated. (b) Replacement of Gly469 (purple) with Glu likely destabilises the OBD by disrupting hydrophobic interactions between β sheets within the OBD.
Given the contribution of DNA ligase IV to cell survival following ionising radiation and the frequency that this modality is used to treat cancer, there has and continues to be interest in the development of DNA ligase IV inhibitors as radiosensitizers, particularly for radioresistant disease that is common in head and neck cancer and colorectal cancer. There is emerging evidence that Wnt signalling contributes to radioresistance in colorectal cancer by increasing expression of DNA ligase IV via β-catenin (96–98). While these studies indicate that a DNA ligase IV inhibitor will reduce the radioresistance of tumours driven by Wnt signalling, it is not clear that this will result in clinical benefit as the inhibitor is also likely to increase radiation sensitivity of adjacent normal tissue.
Conclusion
While there is evidence for significant functional overlap between the three human DNA ligases in proliferating cells, the identification of inherited DNA ligase IV deficiency syndrome and, more recently, DNA ligase I deficiency syndrome demonstrates that these enzymes have unique functions at the organismal level. More work is needed to elucidate the roles and relative contributions of the DNA ligases in different cell types, in particular in stem cells and terminally differentiated cells. The altered expression of DNA ligases in different cancers, together with promising initial results with DNA ligase inhibitors in preclinical cancer models, supports the continued evaluation of DNA ligases as therapeutic targets.
Acknowledgements
We apologise to all colleagues whose work has not been cited because of space limitations.
Funding
Research in the Tomkinson laboratory is supported by National Institutes of Health grants (GM57479, GM47251 ES012512 and CA92584) and by the University of New Mexico Cancer Center, an NCI-designated Comprehensive Cancer Center (CA118100).
Conflict of interest statement: A.E.T. is a co-inventor on patents that cover the use of DNA ligase inhibitors as anti-cancer agents, and altered expression of DSB repair proteins as biomarkers of increased dependence upon alternative non-homologous end joining. The other co-authors have no conflicts.
References
- 1. Ho C. K., Wang L. K., Lima C. D. and Shuman S (2004) Structure and mechanism of RNA ligase. Structure, 12, 327–339. [DOI] [PubMed] [Google Scholar]
- 2. Shuman S. and Schwer B (1995) RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol., 17, 405–410. [DOI] [PubMed] [Google Scholar]
- 3. Ellenberger T. and Tomkinson A. E (2008) Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem., 77, 313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S. H., and Yu X (2018) Human DNA ligase IV is able to use NAD+ as an alternative adenylation donor for DNA ends ligation. Nucleic Acids Res 47, 1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahel I., Rass U., El-Khamisy S. F., Katyal S., Clements P. M., McKinnon P. J., Caldecott K. W. and West S. C (2006) The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature, 443, 713–716. [DOI] [PubMed] [Google Scholar]
- 6. Lakshmipathy U. and Campbell C (1999) The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol., 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lakshmipathy U. and Campbell C (2001) Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res., 29, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao Y., Katyal S., Lee Y., Zhao J., Rehg J. E., Russell H. R. and McKinnon P. J (2011) DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature, 471, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simsek D., Furda A., Gao Y., et al. (2011) Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature, 471, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T. E., Mann M. and Lieber M. R (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 11. Caldecott K. W., McKeown C. K., Tucker J. D., Ljungquist S. and Thompson L. H (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caldecott K. W., Tucker J. D., Stanker L. H. and Thompson L. H (1995) Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caldecott K. W. (2003) Protein-protein interactions during mammalian DNA single-strand break repair. Biochem Soc Trans 31, 247–251 [DOI] [PubMed] [Google Scholar]
- 14. Levin D. S., Bai W., Yao N., O'Donnell M. and Tomkinson A. E (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levin D. S., Vijayakumar S., Liu X., Bermudez V. P., Hurwitz J. and Tomkinson A. E (2004) A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J. Biol. Chem., 279, 55196–55201. [DOI] [PubMed] [Google Scholar]
- 16. Song W., Levin D. S., Varkey J., Post S., Bermudez V. P., Hurwitz J., and Tomkinson A. E (2007) A conserved physical and functional interaction between the cell cycle checkpoint clamp loader and DNA ligase I of eukaryotes. J Biol Chem 282, 22721–22730. [DOI] [PubMed] [Google Scholar]
- 17. Bentley D., Selfridge J., Millar J. K., Samuel K., Hole N., Ansell J. D. and Melton D. W (1996) DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet., 13, 489–491. [DOI] [PubMed] [Google Scholar]
- 18. Bentley D. J., Harrison C., Ketchen A. M., Redhead N. J., Samuel K., Waterfall M., Ansell J. D. and Melton D. W (2002) DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci., 115, 1551–1561. [DOI] [PubMed] [Google Scholar]
- 19. Han L., Masani S., Hsieh C. L., and Yu K (2014) DNA ligase I is not essential for Mammalian cell viability. Cell Rep 7, 316–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arakawa H., Bednar T., Wang M., Paul K., Mladenov E., Bencsik-Theilen A. A. and Iliakis G (2012) Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res., 40, 2599–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liddiard K., Ruis B., Kan Y., Cleal K., Ashelford K. E., Hendrickson E. A. and Baird D. M (2019) DNA Ligase 1 is an essential mediator of sister chromatid telomere fusions in G2 cell cycle phase. Nucleic Acids Res., 47, 2402–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simsek D., and Jasin M (2011) DNA ligase III: a spotty presence in eukaryotes, but an essential function where tested. Cell Cycle 10, 3636–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Chalony C., Hoffschir F., Gauthier L. R., Gross J., Biard D. S., Boussin F. D. and Pennaneach V (2012) Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell. Mol. Life Sci., 69, 2933–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G., Cox L. S., Lane D. P., Abbondandolo A. and Dogliotti E (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 25. Moser J., Kool H., Giakzidis I., Caldecott K., Mullenders L. H. and Fousteri M. I (2007) Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell, 27, 311–323. [DOI] [PubMed] [Google Scholar]
- 26. Okano S., Lan L., Caldecott K. W., Mori T. and Yasui A (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol., 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caldecott K. W. (2007) Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 6, 443–453. [DOI] [PubMed] [Google Scholar]
- 28. Grawunder U., Zimmer D., Fugmann S., Schwarz K. and Lieber M. R (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell, 2, 477–484. [DOI] [PubMed] [Google Scholar]
- 29. Audebert M., Salles B. and Calsou P (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem., 279, 55117–55126. [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Rosidi B., Perrault R., Wang M., Zhang L., Windhofer F. and Iliakis G (2005) DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res., 65, 4020–4030. [DOI] [PubMed] [Google Scholar]
- 31. Simsek D., Brunet E., Wong S. Y., et al. (2011) DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7, e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barnes D. E., Johnston L. H., Kodama K., Tomkinson A. E., Lasko D. D. and Lindahl T (1990) Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA, 87, 6679–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomkinson A. E., Vijayakumar S., Pascal J. M. and Ellenberger T (2006) DNA ligases: structure, reaction mechanism, and function. Chem. Rev., 106, 687–699. [DOI] [PubMed] [Google Scholar]
- 34. Tomkinson A. E., Totty N. F., Ginsburg M. and Lindahl T (1991) Location of the active site for enzyme-adenylate formation in DNA ligases. Proc. Natl. Acad. Sci. USA, 88, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pascal J. M., O'Brien P. J., Tomkinson A. E. and Ellenberger T (2004) Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature, 432, 473–478. [DOI] [PubMed] [Google Scholar]
- 36. Martin I. V. and MacNeill S. A (2002) ATP-dependent DNA ligases. Genome Biol., 3, REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cardoso M. C., Joseph C., Rahn H. P., Reusch R., Nadal-Ginard B. and Leonhardt H (1997) Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol., 139, 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferrari G., Rossi R., Arosio D., Vindigni A., Biamonti G. and Montecucco A (2003) Cell cycle-dependent phosphorylation of human DNA ligase I at the cyclin-dependent kinase sites. J. Biol. Chem., 278, 37761–37767. [DOI] [PubMed] [Google Scholar]
- 39. Frouin I., Montecucco A., Biamonti G., Hübscher U., Spadari S. and Maga G (2002) Cell cycle-dependent dynamic association of cyclin/Cdk complexes with human DNA replication proteins. EMBO J., 21, 2485–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montecucco A., Rossi R., Levin D. S., et al. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J., 17, 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montecucco A., Savini E., Weighardt F., Rossi R., Ciarrocchi G., Villa A. and Biamonti G (1995) The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J., 14, 5379–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rossi R., Villa A., Negri C., Scovassi I., Ciarrocchi G., Biamonti G. and Montecucco A (1999) The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J., 18, 5745–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Levin D. S., McKenna A. E., Motycka T. A., Matsumoto Y. and Tomkinson A. E (2000) Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol., 10, 919–922. [DOI] [PubMed] [Google Scholar]
- 44. Vijayakumar S., Dziegielewska B., Levin D. S., et al. (2009) Phosphorylation of human DNA ligase I regulates its interaction with replication factor C and its participation in DNA replication and DNA repair. Mol. Cell. Biol., 29, 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ferry L., Fournier A., Tsusaka T., et al. (2017) Methylation of DNA Ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol. Cell, 67, 550–565.e5. [DOI] [PubMed] [Google Scholar]
- 46. Barnes D. E., Tomkinson A. E., Lehmann A. R., Webster A. D. and Lindahl T (1992) Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell, 69, 495–503. [DOI] [PubMed] [Google Scholar]
- 47. Webster A. D., Barnes D. E., Arlett C. F., Lehmann A. R. and Lindahl T (1992) Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet, 339, 1508–1509. [DOI] [PubMed] [Google Scholar]
- 48. Maffucci P., Chavez J., Jurkiw T. J., et al. (2018) Biallelic mutations in DNA ligase 1 underlie a spectrum of immune deficiencies. J. Clin. Invest., 128, 5489–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prigent C., Satoh M. S., Daly G., Barnes D. E. and Lindahl T (1994) Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol., 14, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teo I. A., Arlett C. F., Harcourt S. A., Priestley A. and Broughton B. C (1983) Multiple hypersensitivity to mutagens in a cell strain (46BR) derived from a patient with immuno-deficiencies. Mutat. Res., 107, 371–386. [DOI] [PubMed] [Google Scholar]
- 51. Teo I. A., Broughton B. C., Day R. S., James M. R., Karran P., Mayne L. V. and Lehmann A. R (1983) A biochemical defect in the repair of alkylated DNA in cells from an immunodeficient patient (46BR). Carcinogenesis, 4, 559–564. [DOI] [PubMed] [Google Scholar]
- 52. Soza S., Leva V., Vago R., Ferrari G., Mazzini G., Biamonti G. and Montecucco A (2009) DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol. Cell. Biol., 29, 2032–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Henderson L. M., Arlett C. F., Harcourt S. A., Lehmann A. R. and Broughton B. C (1985) Cells from an immunodeficient patient (46BR) with a defect in DNA ligation are hypomutable but hypersensitive to the induction of sister chromatid exchanges. Proc. Natl. Acad. Sci. USA, 82, 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lehmann A. R., Willis A. E., Broughton B. C., James M. R., Steingrimsdottir H., Harcourt S. A., Arlett C. F. and Lindahl T (1988) Relation between the human fibroblast strain 46BR and cell lines representative of Bloom's syndrome. Cancer Res., 48, 6343–6347. [PubMed] [Google Scholar]
- 55. Harrison C., Ketchen A. M., Redhead N. J., O'Sullivan M. J. and Melton D. W (2002) Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res., 62, 4065–4074. [PubMed] [Google Scholar]
- 56. Sun D., Urrabaz R., Nguyen M., Marty J., Stringer S., Cruz E., Medina-Gundrum L. and Weitman S (2001) Elevated expression of DNA ligase I in human cancers. Clin. Cancer Res., 7, 4143–4148. [PubMed] [Google Scholar]
- 57. Montecucco A., Biamonti G., Savini E., Focher F., Spadari S. and Ciarrocchi G (1992) DNA ligase I gene expression during differentiation and cell proliferation. Nucleic Acids Res., 20, 6209–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen J., Tomkinson A. E., Ramos W., Mackey Z. B., Danehower S., Walter C. A., Schultz R. A., Besterman J. M. and Husain I (1995) Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol., 15, 5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei Y. F., Robins P., Carter K., et al. (1995) Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol., 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mackey Z. B., Ramos W., Levin D. S., Walter C. A., McCarrey J. R. and Tomkinson A. E (1997) An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol., 17, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lakshmipathy U. and Campbell C (2000) Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res., 28, 3880–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nash R. A., Caldecott K. W., Barnes D. E. and Lindahl T (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 63. Okano S., Lan L., Tomkinson A. E. and Yasui A (2005) Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res., 33, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chiang S. C., Carroll J. and El-Khamisy S. F (2010) TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle, 9, 588–595. [DOI] [PubMed] [Google Scholar]
- 65. Cotner-Gohara E., Kim I. K., Tomkinson A. E. and Ellenberger T (2008) Two DNA-binding and nick recognition modules in human DNA ligase III. J. Biol. Chem., 283, 10764–10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mackey Z. B., Niedergang C., Murcia J. M., Leppard J., Au K., Chen J., de Murcia G. and Tomkinson A. E (1999) DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem., 274, 21679–21687. [DOI] [PubMed] [Google Scholar]
- 67. Cotner-Gohara E., Kim I. K., Hammel M., Tainer J. A., Tomkinson A. E. and Ellenberger T (2010) Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry, 49, 6165–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kukshal V., Kim I. K., Hura G. L., Tomkinson A. E., Tainer J. A. and Ellenberger T (2015) Human DNA ligase III bridges two DNA ends to promote specific intermolecular DNA end joining. Nucleic Acids Res., 43, 7021–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yoon G. and Caldecott K. W (2018) Nonsyndromic cerebellar ataxias associated with disorders of DNA single-strand break repair. Handb. Clin. Neurol., 155, 105–115. [DOI] [PubMed] [Google Scholar]
- 70. Canugovi C., Shamanna R. A., Croteau D. L. and Bohr V. A (2014) Base excision DNA repair levels in mitochondrial lysates of Alzheimer's disease. Neurobiol. Aging, 35, 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Akbari M., Keijzers G., Maynard S., Scheibye-Knudsen M., Desler C., Hickson I. D. and Bohr V. A (2014) Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst)., 16, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tobin L. A., Robert C., Rapoport A. P., Gojo I., Baer M. R., Tomkinson A. E. and Rassool F. V (2013) Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene, 32, 1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tobin L. A., Robert C., Nagaria P., et al. (2012) Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol. Cancer Res., 10, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Newman E. A., Lu F., Bashllari D., Wang L., Opipari A. W. and Castle V. P (2015) Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Mol. Cancer Res., 13, 470–482. [DOI] [PubMed] [Google Scholar]
- 75. Muvarak N., Kelley S., Robert C., Baer M. R., Perrotti D., Gambacorti-Passerini C., Civin C., Scheibner K. and Rassool F. V (2015) c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in tyrosine kinase-activated leukemias. Mol. Cancer Res., 13, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sallmyr A., Matsumoto Y., Roginskaya V., Van Houten B. and Tomkinson A. E (2016) Inhibiting mitochondrial DNA ligase IIIα activates caspase 1-dependent apoptosis in cancer cells. Cancer Res., 76, 5431–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grawunder U., Zimmer D., Kulesza P. and Lieber M. R (1998) Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem., 273, 24708–24714. [DOI] [PubMed] [Google Scholar]
- 78. Grawunder U., Zimmer D. and Lieber M. R (1998) DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr. Biol., 8, 873–876. [DOI] [PubMed] [Google Scholar]
- 79. Pannunzio N. R., Watanabe G. and Lieber M. R (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem., 293, 10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. DeFazio L. G., Stansel R. M., Griffith J. D. and Chu G (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J., 21, 3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hammel M., Yu Y., Mahaney B. L., et al. (2010) Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem., 285, 1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cottarel J., Frit P., Bombarde O., et al. (2013) A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol., 200, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hammel M., Yu Y., Radhakrishnan S. K., et al. (2016) An intrinsically disordered APLF Links Ku, DNA-PKcs, and XRCC4-DNA Ligase IV in an extended flexible non-homologous end joining complex. J Biol Chem 291, 26987–27006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang Y., Lamarche B. J. and Tsai M. D (2007) Human DNA ligase IV and the ligase IV/XRCC4 complex: analysis of nick ligation fidelity. Biochemistry, 46, 4962–4976. [DOI] [PubMed] [Google Scholar]
- 85. Riballo E., Woodbine L., Stiff T., Walker S. A., Goodarzi A. A. and Jeggo P. A (2009) XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res., 37, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kaminski A. M., Tumbale P. P., Schellenberg M. J., Williams R. S., Williams J. G., Kunkel T. A., Pedersen L. C. and Bebenek K (2018) Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis. Nat. Commun., 9, 2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Frank K. M., Sharpless N. E., Gao Y., et al. (2000) DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell, 5, 993–1002. [DOI] [PubMed] [Google Scholar]
- 88. Frank K. M., Sekiguchi J. M., Seidl K. J., Swat W., Rathbun G. A., Cheng H. L., Davidson L., Kangaloo L. and Alt F. W (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature, 396, 173–177. [DOI] [PubMed] [Google Scholar]
- 89. Barnes D. E., Stamp G., Rosewell I., Denzel A., and Lindahl T (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol 8, 1395–1398 [DOI] [PubMed] [Google Scholar]
- 90. Altmann T. and Gennery A. R (2016) DNA ligase IV syndrome; a review. Orphanet J. Rare Dis., 11, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Riballo E., Critchlow S. E., Teo S. H., et al. (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol., 9, 699–702. [DOI] [PubMed] [Google Scholar]
- 92. Buck D., Moshous D., de Chasseval R., et al. (2006) Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur. J. Immunol., 36, 224–235. [DOI] [PubMed] [Google Scholar]
- 93. Toita N., Hatano N., Ono S., et al. (2007) Epstein-Barr virus-associated B-cell lymphoma in a patient with DNA ligase IV (LIG4) syndrome. Am. J. Med. Genet. A, 143A, 742–745. [DOI] [PubMed] [Google Scholar]
- 94. O'Driscoll M., Cerosaletti K. M., Girard P. M., et al. (2001) DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell, 8, 1175–1185. [DOI] [PubMed] [Google Scholar]
- 95. O'Driscoll M., Gennery A. R., Seidel J., Concannon P., and Jeggo P. A (2004) An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 3, 1227–1235 [DOI] [PubMed] [Google Scholar]
- 96. Jun S., Jung Y. S., Suh H. N., et al. (2016) LIG4 mediates Wnt signalling-induced radioresistance. Nat. Commun., 7, 10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Woodward W. A., Chen M. S., Behbod F., Alfaro M. P., Buchholz T. A. and Rosen J. M (2007) WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA, 104, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen M. S., Woodward W. A., Behbod F., Peddibhotla S., Alfaro M. P., Buchholz T. A. and Rosen J. M (2007) Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J. Cell Sci., 120, 468–477. [DOI] [PubMed] [Google Scholar]