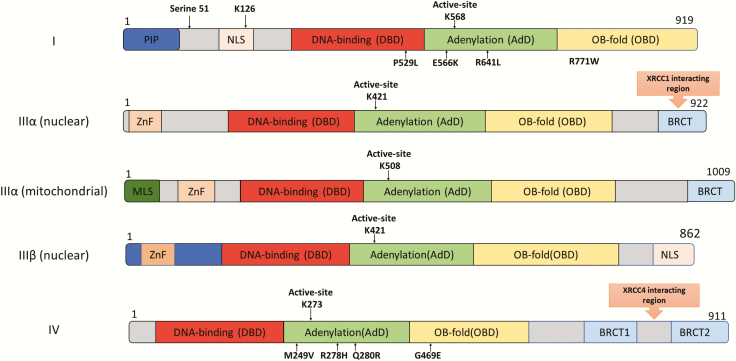
Fig. 2.
Domain organisation of the DNA ligases encoded by the human LIG1, LIG3 and LIG4 genes. The Adenylation Domain (AdD, green) and Oligonucleotide/Oligosaccharide Binding-fold Domain (OBD, yellow) domains comprise the catalytic core that contains the key active site lysine residue. The less conserved DNA Binding Domain (DBD, red) is N-terminal to this core. The non-catalytic N-terminal region of DNA ligase I contains PCNA interaction peptide, PIP (blue) and nuclear localisation signal (beige). The three isoforms of DNA ligase III have an N-terminal Zinc finger domain (light orange: ZnF). Mitochondrial DNA ligase IIIα has a mitochondrial localisation signal, MLS (dark green) at the N-terminus. DNA ligase IIIα and DNA ligase IV contain one and two C-terminal BRCT domains (blue), respectively. The DNA ligase IIIα BRCT domain is required for interaction with XRCC1 and whereas XRCC4 interacts with the region between the DNA ligase IV BRCT domains. Amino acid substitutions identified in DNA ligase deficiency syndromes are indicated below the DNA ligase polypeptides.