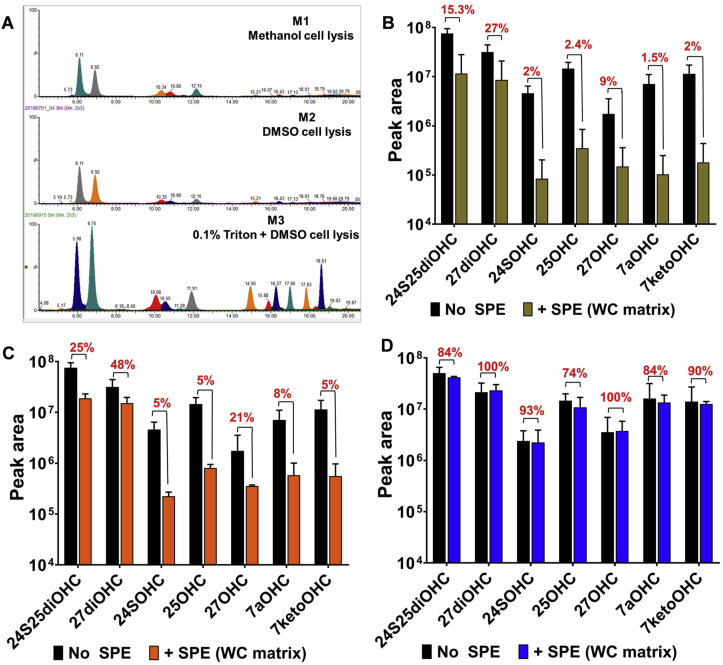
Fig. 2.
Comparison of oxysterol recovery and matrix effects from three methods. (A) Three methods; M1 (methanol (MeOH) cell lysis and SPE oxysterol extraction, M2 dimethylsulfoxide (DMSO) cell lysis, methanol:dichloromethane (MeOH:DCM) lipid isolation and SPE oxysterol extraction, M3 (0.1% Triton X-100 + DMSO cell lysis, MeOH:DCM lipid extraction and SPE oxysterol extraction) were compared. Authentic standards were prepared in 40% methanol 0.1% formic acid (without cellular matrix and no SPE extraction) and used as the control (black bars). M3 improved chromatographic resolution of 10ng authentic oxysterol standards mixed with whole cell lysate as the matrix and isolated using the methodology described in Fig. 1 (B), (C) and (D) compares the recovery (ratio of +SPE peak area/No SPE peak area) expressed as % for the authentic standards in matrix using extraction methods M1 [29], M2 [38] and M3-developed in this work. M3 showed >70% recovery for all nine oxysterols. Measurements were done using standards at 1ng.μl-1 and are mean ± S.D. of 3–4 independent replicates.