Abstract
Background
Alopecia areata (AA) is considered a highly heritable, T-cell-mediated autoimmune disease of the hair follicle. However, no convincing susceptibility gene has yet been pinpointed in the major histocompatibility complex (MHC), a genome region known to be associated with AA as compared to other regions.
Methods
We engineered mice carrying AA risk allele identified by haplotype sequencing for the MHC region using allele-specific genome editing with the CRISPR/Cas9 system. Finally, we performed functional evaluations in the mice and AA patients with and without the risk allele.
Findings
We identified a variant (rs142986308, p.Arg587Trp) in the coiled-coil alpha-helical rod protein 1 (CCHCR1) gene as the only non-synonymous variant in the AA risk haplotype. Furthermore, mice engineered to carry the risk allele displayed a hair loss phenotype. Transcriptomics further identified CCHCR1 as a novel component interacting with hair cortex keratin in hair shafts. Both, these alopecic mice and AA patients with the risk allele displayed morphologically impaired hair and comparable differential expression of hair-related genes, including hair keratin and keratin-associated proteins (KRTAPs).
Interpretation
Our results implicate CCHCR1 with the risk allele in a previously unidentified subtype of AA based on aberrant keratinization in addition to autoimmune events.
Funding
This work was supported by JSPS KAKENHI (JP16K10177) and the NIHR UCLH Biomedical Research center (BRC84/CN/SB/5984).
Keywords: Alopecia areata, CRISPR/Cas9, MHC, Haplotype, Association, Sequencing
Research in context.
Evidence before this study
Alopecia areata (AA) is classified as a multi-factorial autoimmune disease. It is generally assumed that the immunological pathogenesis of AA requires infiltration of a subset of T cells within surrounding AA hair follicles. However, lymphocytic infiltration was not detected in all AA skin specimens, and inhibition of immunological pathway though pharmacological inhibitors was not effective in all AA patients. Therefore, AA appears to display a more complex etiology than previously thought but similar to other common diseases. Moreover, no studies to date have identified any AA-susceptibility variants supported by functional evidence, although previous genome-wide association studies (GWASs) identified susceptibility loci including the major histocompatibility complex (MHC) displaying strongest genetic association with many autoimmune diseases.
Added value of this study
We have discovered a nonsynonymous variant in the MHC delineating AA susceptibility in the CCHCR1 (coiled-coil alpha-helical rod protein 1) coding a novel component of hair shafts. In addition, the present results demonstrate that mice carrying the amino acid substitution display a hair loss phenotype. We further identify keratin abnormalities on the hair shaft and comparative differential expression of hair-related keratin genes not only in the alopecic mice but also in hair follicles from AA patients with the risk variant. Thus, our study identifies a novel AA susceptibility variant validated by functional analysis.
Implications of all the available evidence
Results of the present genetic and functional analyses identified keratin abnormalities on the hair shaft as an additional pathway of AA pathogenesis, thus resulting in a previously unidentified subtype of AA. The alopecic mice engineered in this study represent a resource for future research regarding this new AA subtype as well as AA in general. Specifically, the present results provide important information for developing genotype-based diagnostics and drug-based therapeutics, in addition to the well-studied immunological pathway. Moreover, our experimental findings present a rare example of a non-mendelian common disease variant demonstrating biological functions related to disease phenotypes.
Alt-text: Unlabelled box
1. Introduction
Alopecia areata (AA) is a multi-factorial diseases defined by focal or universal hair loss, and is classified as an autoimmune disease depending on its clinical features [1] including infiltration of CD8+NKG2D+ T cells within surrounding AA hair follicles [2]. Life time risk of AA is estimated to be 2% in the United States [3], while a twin study suggested a 55% concordance rate in identical twins with a significant occurrence of AA in families [4]. In addition, the prevalence rate of AA in families has been shown to be higher than that in the general public, though the rate varied in each study and population examined [5], [6], [7], [8], [9]. Environmental factors such as infection and psychological stress may also play important roles [5]. AA is driven by cytotoxic T lymphocytes and was found to be reversible by Janus kinase (JAK) inhibition in clinical treatment [2]. However, the peribulbar lymphocyte infiltration was not detected in skin specimens of all AA patients [10], and JAK inhibitors were not effective for all AA patients [11].
Previous genome-wide association studies (GWAS) have implicated a number of immune and non-immune loci in the etiology of AA [12], [13], [14], though none has yet been demonstrated to be causative for the disease and none has been functionally validated to be involved in AA pathogenesis. Alleles of the human leukocyte antigen (HLA) genes within the major histocompatibility complex (MHC) on chromosome 6p21.3 have so far shown the strongest associations with AA across different ethnic groups [12], [13], [14]. The largest reported genome-wide meta-analysis of AA demonstrated HLA-DRβ1 as a key etiologic driver [13]. However, the strongest associations with AA have not been supported by functional evidence.
The genetic architecture of the MHC region shows that multiple haplotypes with the highest degree of diversity are often maintained in a population by balancing selection, and that positive selection can occasionally generate long-range haplotypes [15,16]. The strong linkage disequilibrium (LD) observed in such haplotypes can mask the ability to discriminate between a bona fide variant associated with disease and a variant influenced by LD. This limitation can be addressed by analysis of microsatellites that have higher mutation rates than SNPs, thus leading to breakup of apparently invariant SNP haplotypes into lower frequency haplotypes for functional analysis [17]. Analysis of multi-allelic microsatellites may therefore be an effective strategy for identifying rare disease-associated haplotypes in the MHC.
With this background in mind, we implemented a 4-step study design. First, we performed association analysis using microsatellites for the entire MHC region with AA patients and healthy controls to identify risk haplotypes associated with AA. Second, we sequenced representative risk and control haplotypes to identify variants that were present only in identical risk haplotypes based on all of the variants detected. Third, for the confirmation of the AA susceptibility allele we engineered mice carrying the human risk allele using allele-specific genome editing with the CRISPR/Cas9 system and performed morphologically observations and functional evaluations. Finally, we also investigated subjects of AA patients with and without the risk allele.
2. Materials and Methods
2.1. Patients and controls for association and sequencing analysis
Upon approval of the experimental procedures from the relevant ethical committees of Juntendo University (reference number:2,013,097) and Tokai University (reference number: 131–07), we obtained informed consent from all unrelated AA and healthy individuals prior to collection of DNA samples. A total of 171 individuals affected with AA (MAA-multiple alopecia areata: 115, AT-alopecia totalis: 18, AU-alopecia universalis: 38) and 560 unrelated individuals of Japanese origin participated in this study. All cases were diagnosed and treated at a Juntendo University Hospital in Japan. DNA was extracted using a QIAamp DNA blood kit (QIAGEN, Hilden, Germany) under standardized conditions to prevent variations in DNA quality. For additional quality control, we used 0.8% agarose gel electrophoresis to check for DNA degradation and/or RNA contamination and performed optical density measurements to check for protein contamination. The final DNA concentration was determined with 3 successive measurements using a PicoGreen fluorescence assay (Molecular Probes, Thermo Fisher Scientific, Inc., Waltham, MA, USA).
2.2. Microsatellite genotyping
We selected 22 microsatellites spanning 2.44 Mbp (from HLA-E to PSMB9 gene) in the HLA region (Supplementary Table 1) harbouring the HLA class I, II, class III regions, and genotyped all patients and control subjects. Forward primers of the primer sets to amplify microsatellites were labeled by 5′ fluorescent FAM. Oligonucleotides were obtained from Greiner Bio-one. PCR and fragment analyses were performed with capillary electrophoresis using an Applied Biosystems 3730 Genetic Analyzer. Allele assignment was determined with GeneMapper Software (Thermo Fisher Scientific) and conducted as previously described [18]. Fragment sizes were assigned to allele names in the corresponding microsatellites. In the MICA locus, 5 MICA polymorphisms (A4, A5, A5.1, A6, A9) were determined based on the number of alanine (GCT) repeats. The A5.1 allele contained 5 GCT repeats, plus 1 extra guanine nucleotide (GCT)2G(GCT)3.
2.3. HLA-C locus genotyping
A LABType® SSO typing test produced by ONE LAMBDA (Inc., Canoga Park, CA) was utilized. This product is based on the reverse SSO method for use with a suspension array platform with microspheres as a solid support to immobilize oligonucleotide probes. Target DNA is amplified by PCR, then hybridized to the bead probe array, followed by flow analysis using a LABScan™ 100 flow analyzer (ONE LAMBDA). HLA-C locus genotype data from 156 AA patients and 560 controls obtained in our previous study were used and we also genotyped an additional 15 AA cases as part of the present study [19].
2.4. Genomic library construction and sequencing
For HLA region capture and sequencing, genomic DNA (2 μg) was sheared to approximately 500 bp in size using a Covaris Acoustic Adaptor. Genomic libraries were prepared using a TruSeq DNA Sample Preparation kit. v.2 (Illumina, San Diego, CA, USA) following the manufacturer's instructions, which involved size selection of DNA fragments of 550–650 bp in length on 2% agarose gels. HLA region enrichment was performed with an adaptor-ligated DNA sample library using the SeqCap EZ Choice Library Human MHC Design system (Roche NimbleGen, Madison, WI, USA) [20], according to the manufacturer's instructions. To quantify and verify the genomic libraries, all samples were analyzed with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) using an Agilent DNA 1000 kit prior to sequencing (Supplementary Fig. 1). Sequence analysis was performed with the Illumina Genome Analyzer IIx platform, using a paired-end sequencing protocol (2 × 100 bp). We sequenced 5 risk and 7 non-risk haplotypes to a mean depth of 249 reads, covering 95.2% (mean) of the 4.97-Mb MHC region (chr6:28,477,797–33,451,433, hg19) with at least 10 reads (Supplementary Table 2).
2.5. Next generation sequencing (NGS) data analysis
Fastx-toolkit, v. 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/index.html), was used for quality control of the sequencing reads. Reads that passed quality control were mapped to the human reference genome (UCSC Genome Browser assembly GRCh37/hg19, http://genome.ucsc.edu/) using Burrows-Wheeler Aligner (BWA), v. 0.5.9, with the default parameters [21]. After alignment, Sequence Alignment/Map (SAMtools), v. 0.1.17, was used to convert the .sam to .bam files [22], and potential PCR duplicates were flagged with Picard MarkDuplicates (v. 1.88; http://picard.sourceforge.net/). A Genome Analysis Toolkit (GATK, v. 2.2–8) was used to perform local realignment, map quality score recalibration, and variant detection [23]. SNVs and indels were then annotated for functional consequences at the gene and protein sequence levels using ANNOVAR [24]. Finally, we manually checked the raw sequencing data using Tablet, a sequence assembly visualization tool (Supplementary Fig. 2) [25].
2.6. Variant discovery and genotyping of CCHCR1 by Sanger sequencing
Coding exons of CCHCR1 were sequenced using PCR-based capillary Sanger sequencing. Oligonucleotides were purchased from Greiner Bio-One (Supplementary Table 3). PCR was performed in a reaction volume of 10 μl containing 5 ng of genomic DNA, 0.2 U of KOD FX Neo (TOYOBO Life Science, Osaka, Japan), 5 μl of 2 × PCR Buffer, 2 μl of dNTP (2 mM each), and 0.2 μM (final concentration) of each of the primers. The thermal cycling profile was as follows: initial denaturation at 94 °C for 2 min and 35 rounds of amplification at 98 °C for 10 s, then 59–65 °C (depending on primer set; see Supplementary Table 3) for 30 s and 68 °C for 1 min. PCR products were purified using an AMPure XP (Beckman Coulter, Fullerton, CA, USA), according to the manufacturer's protocol. Purification and sequencing of the PCR products were carried out using a BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific) and BigDye XTerminator Purification Kit (Thermo Fisher Scientific), following the manufacturer's instructions. Automated electrophoresis was performed with an ABI PRISM 3730 Genetic Analyzer (Thermo Fisher Scientific). Sequencing data were analyzed using Sequencher (v. 5.1, Gene Codes Corporation, Ann Arbor, MI, USA). Genomic coordinates for all variants were called using the UCSC Genome Browser assembly GRCh37/hg19 (http://genome.ucsc.edu/).
2.7. Statistical analysis
Logistic regression models were used to assess the genetic effects of multi-allelic loci, SNVs, and AA risk haplotypes. Comparisons of genotype and haplotype frequency differences were done by regression analysis for log-additive models [26]. Unadjusted odds ratio (OR) and 95% confidence intervals (95% CI) were calculated. Analysis was carried out using the SNPassoc R library [26]. For these association analyses, we used Bonferroni-corrected values to account for the problem of multiple testing to a threshold P value of 1.98 × 10−04, after accounting for multiple testing of 252 alleles in 23 multi-allelic loci for the first microsatellite analysis. An exact P value test of Hardy–Weinberg proportion and evaluation of LD (linkage disequilibrium) for multi-allelic loci were simulated by the Markov chain method within Genepop [27]. To evaluate the degree of LD for bi-allelic loci (SNVs) we used Haploview 4.2 [28]. To estimate haplotypes for SNVs and multi-allelic loci, we used PHASE v2.1.1 [29]. For EHH analysis, haplotype data were generated from 22 nonsynonymous SNVs, 2 stop gain SNVs, and 19 multi-allelic loci with fastPHASE v1.2 [30]. Moreover, for this estimation, each multi-allelic locus was regarded as a SNV [15]. Thus, we were able to extract the allele demonstrating LD (D' ≥0.5) for the AA risk allele (rs142986308: T allele) from all alleles in each locus, if such an allele was detected (Supplementary Fig. 3), and then merged it with the other alleles. EHH was calculated using the “rehh” package [31]. To estimate the power of this study design to detect associated loci with AA, we performed statistical power calculations using the Genetic Power Calculator web application (http://pngu.mgh.harvard.edu/∼purcell/gpc/) assuming a type I error of alpha (a) = 0.05 [32]. The statistical power for a significance level of a = 0.05 in our sample was calculated for the D6S2930 locus under several assumptions, as follows: a) high risk allele frequency of 0.144 in the general population, b) prevalence for AA of 0.001, c) heterozygote genotype relative risk of 1.76, d) homozygote genotype relative risk of 4.66, and e) a control group of subjects comprised of unselected individuals from the general population not screened for AA. Based on these conditions, the statistical power for the D6S2930 locus was calculated to be 0.781.
2.8. Prediction of coiled-coil domain and structural analyses
Multiple sequence alignment was performed with CLUSTALW (http://www.genome.jp/). To predict coiled-coil domains within the amino acid sequence of CCHCR1, we used 2 different programs, COILS [33] (http://embnet.vital-it.ch/software/COILS_form.html) and Paircoil2 (http://groups.csail.mit.edu/cb/paircoil2/),[34,35] with the following recommended default settings: COILS v2.2 with a window size of 28 and the MTIDK table, and Paircoil2 with a window size of 28 and P-score cutoff of 0.025.
The amino acid sequence for CCHCR1 (GenBank accession: NP_061925.2) was obtained from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov) and used as a target for homology modeling. Following PSI-BLAST searches of protein databank (PDB) sequence entries (http://www.rcsb.org/pdb/) using the CCHCR1 sequence [36], the crystal structure of the human lamin-B1 coil 2 segment [PDB ID: 3TYY] was selected as the best template for homology modeling [37]. The partial 3TYY structure was optimized and utilized as a template to generate 50 homology models using the Build Homology Models protocol. Probability density function total and discrete optimized potential energy scores were then used to select the optimal model. Protein stability of the mutants was calculated using the Calculate Mutation Energy (Stability) protocol. All molecular modeling and simulations were performed with Discovery Studio, v. 4.1, from BIOVIA (Accelrys Inc., San Diego, USA) using the default parameter setting.
2.9. Genome editing by CRISPR/Cas9 in mouse embryos
To generate mice carrying the p.Arg587Trp disease-associated missense variant, we edited the Cchcr1 codon sequence of amino acid 591 in mice using the following protocol (Supplementary Fig. 4). Cas9 mRNA was prepared using a pBGK plasmid, as previously described [38]. The plasmid was linearized with XbaI and used as the template for in vitro transcription with an mMESSAGE mMACHINE T7 ULTRA kit (Ambion, Foster City, CA, USA). Single guide RNAs (sgRNAs) (guide1: GCTGTGTCAGCTCCTGACGG[AGG], guide2: GGAAGCTGCCAGCCTCCGTC[AGG]) were designed using CRISPR design (CRISPR.mit.edu). The templates for sgRNA synthesis were PCR amplified with primer sets (5′-TAATACGACTCACTATAGGGCTGTGTCAGCTCCTGACGGGTTTTAGAGCTAGAAATAGCAAG-3′ / 5′-AAAAAAAGCACCGACTCGG-3′ for sgRNA1, 5′-TAATACGACTCACTATAGGGGAAGCTGCCAGCCTCCGTCGTTTTAGAGCTAGAAATAGCAAG-3′ / 5′-AAAAAAAGCACCGACTCGG-3′ for sgRNA2) using pUC57-sgRNA vector (Addgene number: #51,132) as a template. Then, 400 ng of gel-purified PCR products were subjected to RNA synthesis with a MEGAshortscript T7 Kit (Ambion), according to the manufacturer's instructions [39]. Both Cas9 mRNA and sgRNA were purified with a MEGAclear kit (Ambion) and filtered by passing through an Ultrafree-MC filter (HV; 0.45 µm pore size; Millipore, Billerica, MA) to avoid clogging during microinjection. The single-stranded oligodeoxynucleotide (ssODN) (5′-CAGCAGTTGGAGGCAGCACGTCGGGGCCAGCAGGAGAGCACGGAGGAAGCTGCCAGCCTCtGgCAGGAGCTGACACAGCAGCAGGAAATCTACGGGCAAGGTGTGGGGGCGTGGCGGTGTGTG-3′) was synthesized by IDT (Coralville, IA, USA). C57BL/6 N strain mouse zygotes were obtained using in vitro fertilization. One-cell stage fertilized mouse embryos were injected with 10 ng/µl of Cas9 mRNA, 10 ng/µl of sgRNA, and 20 ng/µl of ssODN. Injected eggs were cultured overnight in KSOM medium and the resulting two-cell embryos were transferred into the oviducts of pseudo-pregnant ICR females, as previously described [39].
2.10. Mice
C57BL/6NJcl mice were purchased from CLEA (Shizuoka, Japan) and maintained under specific pathogen-free conditions. Wild-type mice (8- to 10-month-old females and males) were used as control group and/or for calibration. This study used 27 mice (8- to 12-month-old females and males) carried the mutation generating amino-acid substitution. Randomization and blinding tests were not performed in this study. All animal procedures were done according to protocols approved by the Institutional Animal Care and Use Committee of Tokai University. We registered the allele name of Cchcr1em1Aoka (MGI:6,394,064) for mice established in this study based on the protocol of the International Mouse Genetic Nomenclature Committee (http://www.informatics.jax.org/).
2.11. Genotyping allele of target locus for generated mice with alkaline lysis method
Mouse tissue obtained by ear punch was added to 180 μl of a 50-mM NaOH solution and incubated at 95 °C for 10 min. The lysate for PCR was obtained by neutralizing with 20 μl of 1 M Tris–HCl (pH 8.0) and centrifugation. PCR was performed in a reaction volume of 10 μl containing 1 μl of lysate, 0.2 U of KOD FX Neo (TOYOBO), 5 μl of 2 × PCR buffer, 2 μl of dNTP (2 mM each), and 0.2 μM (final concentration) of each primer. Forward (AGCTGAGTGCCCACCTGAT) and reverse (TGTGTCTCAGTGCTGCCTTC) primers were used for sequencing (Greiner Bio-one). The thermal cycling profile was as follows: initial denaturation at 94 °C for 2 min, then 35 rounds of amplification at 98 °C for 10 s and 60 °C for 25 s. The protocol for sequencing was performed as previously described in as above in ‘2.7, Variant discovery and genotyping of CCHCR1 by Sanger sequencing’.
2.12. Morphology of hair shafts of mice
Hairs were plucked from mice and analyzed by SEM using a JSM 6510LV (Jeol Co., Tokyo, Japan).
2.13. Isolation of mouse skin RNA
Total RNA was isolated from sections of mouse skin using ISOGEN (Nippon Gene, Tokyo, Japan), according to the manufacturer's protocol, and treated twice with TURBO DNase (Ambion) to eliminate contaminating DNA. RNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific) and the quality of the extracted RNA was evaluated with a Bioanalyzer 2100 (Agilent Technologies).
2.14. Microarray analysis of mouse skin RNA
Fluorescent cRNA synthesis derived from skin RNAs was performed using a Low RNA Input Linear Amplification kit (Agilent Technology) and subjected to DNA microarray analysis with single-color microarray-based gene-expression analysis (SurePrint G3 Mouse GE, v. 2.0, 8 × 60 K, Agilent Technology). All procedures were performed according to the manufacturer's instructions. Data from samples that passed the QC parameters were subjected to 75th percentile normalization (Agilent Feature Extraction 11.5.1.1, Agilent Technologies) and analyzed using Genespring GX (version 12, Agilent Technologies).
2.15. Quantitative PCR of mouse skin RNA
cDNAs were synthesized using 1 μg of total RNA in a 20-μl total volume using SuperScript® VILO™ MasterMix (Thermo Fisher Scientific) and random hexamers. Quantitative PCR was performed using a StepOnePlus™ Real-Time PCR System, TaqMan® Universal Master Mix, and TaqMan gene expression assay (Thermo Fisher Scientific), according to the manufacturer's protocol. The primer-probe sets were as follows: Mm00652053_g1 (Krt34), Mm02345064_m1 (Krt73), Mm04208593_s1 (Krtap3-3), Mm04336701_s1 (Krtap16-1), Mm00478075_m1 (Padi3), Mm01214103_g1 (S100a3), Mm00461542_m1 (Cchcr1), and Mm99999915_g1 (Gapdh) (Thermo Fisher Scientific). All PCR reactions were performed in triplicate. Relative quantification of gene expression was performed using the 2−ΔΔCt method [40]. Fold change values were calculated using Gapdh as the internal control and a dorsal skin sample from a wild-type mouse was used as a calibrator. Significance values were calculated between the Cchcr1- and wild-type mice using Welch's t-test (unpaired, two-tailed).
2.16. Immunohistochemistry
Anti-CCHCR1 (rabbit polyclonal) and anti-hair cortex cytokeratin antibodies (mouse monoclonal [AE13]) were obtained from Novus Biologicals (NBP2-29,926) (Littleton, CO, USA) and Abcam (ab16113) (Cambridge, MA, USA), respectively. Rabbit IgG (rabbit polyclonal) and Mouse IgG1,k (mouse monoclonal [MG1-45]) for these isotype controls were obtained from MBL (PM035) (Nagoya, Japan) and BioLegend (401,407) (San Diego, CA, USA), respectively. This CCHCR1 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 599–627 amino acids from the central region of human CCHCR1. The homology of the 29 peptides between human and mouse is 97% (28/29). Anti-CD4 (rabbit monoclonal [EPR19514]) and anti-CD8 antibodies (rabbit monoclonal [EPR21769]) were obtained from Abcam (Cambridge, MA, USA).
Deparaffinized skin sections for anti-CCHCR1 and anti-hair cortex cytokeratin antibodies staining were boiled in 10 mM of citrate buffer (pH 5.0) for antigen unmasking. Sections were incubated in Blocking One Histo (Nacarai tesque, Kyoto, Japan) for 30 min at room temperature, then incubated with primary antibodies [anti-CCHCR1 (1:100), anti-hair cortex Cytokeratin (1:200)] in PBS containing 5% Blocking One Histo and 0.05% Triton-X 100 overnight at 4°C. Sections were then washed and incubated with secondary antibodies [anti-mouse IgG-Alexa488 (1:500), anti-rabbit IgG-Alexa594 (1:500)] for 2 h at room temperature. Controls for all immunostaining were simultaneously performed by omitting the primary antibody. Sections were cover-slipped using Vectashield with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Vector Laboratories, Burlingame, CA, USA) for nuclei counterstaining and analyzed with Keyence BZ X-700 (Keyence, Tokyo, Japan).
Deparaffinized skin sections for anti-CD4 and anti-CD8 antibodies staining were boiled in Tris-EDTA buffer (pH 9.0) for antigen unmasking. Sections were incubated in Blocking One Histo for 30 min at room temperature, then incubated with primary antibodies [anti-CD4 (1:2000), anti-CD8 (1:2000)] in PBS containing 5% Blocking One Histo and 0.05% Tween 20 overnight at 4°C. Sections were then washed and incubated with peroxidase (PO)-conjugated goat anti-rabbit IgG (Histofine Simple Stain Mouse MAX-PO, Nichirei Bioscience, Tokyo, Japan) for 2 h at room temperature. Controls for immunostaining were simultaneously performed using rabbit monoclonal [DA1E] antibody IgG obtained from Cell Signaling Technology (Tokyo, Japan) as isotype control. The ImmPACT DAB substrate Kit (Vector Laboratories, Burlingame, CA, USA) was used for staining with Diaminobenzidine (DAB).
2.17. PLA assay
PLA assay was conducted using Duolink® Proximity Ligation Assay (PLA) (SIGMA). Anti-CCHCR1 and anti-hair cortex cytokeratin antibodies described in Immunohistochemistry section were used as primary antibodies. The 2 primary antibodies raised in different species were incubated with the samples to detect two specific protein targets, were followed by the incubation with secondary antibodies coupled with oligonucleotides (PLA probes). Then, connector oligos joined the PLA probes were ligated. The resulting closed, circular DNA template were amplified by DNA polymerase. Complementary detection oligos coupled to fluorochromes hybridized to repeating sequences in the amplicons. PLA signals were detected by BZ-X800 (Keyence).
2.18. Hair follicles from AA patients
Upon approval of the experimental procedures from the relevant ethical committees of Juntendo University (reference number:2,018,067), we obtained informed consent from all AA patients prior to collection of hair samples. A total of 3 individuals affected with AA of Japanese origin participated in this study. All cases were diagnosed and treated at a Juntendo University Hospital in Japan. Hairs around hair loss areas were plucked from AA patients.
2.19. Morphology of hair shafts of AA patients
Hairs of AA patients were analyzed by SEM using a S4800 (Hitachi, Tokyo, Japan).
2.20. Isolation of human hair follicle RNA
To extract RNA, we use 10 hairs of unstaged follicles were plucked from the scalp of 3 AA patients.
Hairs were rinsed out contaminants by ethanol, cut off by sterilized scissors to be about 5 mm long including hair root, and immediately frozen in a 1.5-ml sterile centrifuge tube by liquid nitrogen. Total RNA derived from hair root was extracted by RNeasy micro kit (QIAGEN, Hilden, Germany). Tissue disruption was performed by sample homogenizer SH-100 (Kurabo Industries, Ltd., Osaka, Japan).
2.21. Microarray analysis of human hair follicle RNA
10 ng of total RNA from human hair follicle was processed for use on the microarray using the GeneChip WT pico Reagent Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The resultant single-strand cDNA was fragmented and labeled with biotin, then hybridized to the GeneChip Human Gene 2.0 ST Array. The arrays were washed, stained and scanned using the Affymetrix 450 Fluidics Station and GeneChip Scanner 3000 7 G (Thermo Fisher Scientific) according to the manufacturer's recommendations. Expression values were generated using Expression Console software (version 1.3 Thermo Fisher Scientific) with default robust multichip analysis parameters and evaluated by Genespring GX (version 12, Agilent Technologies).
2.22. Quantitative PCR of human hair follicle RNA
cDNAs were synthesized using 400 ng of total RNA in a 20-μl total volume using SuperScript® VILO™ MasterMix and random hexamers. Quantitative PCR was performed using a ViiA™7 Real-Time PCR System, TaqMan® Universal Master Mix II, and TaqMan gene expression assay (Thermo Fisher Scientific), according to the manufacturer's protocol. The primer-probe sets were as follows: Hs03037438_s1 (KRTAP5-5), Hs00799866_s1 (KRTAP6-1), Hs00955092_m1 (KRT25), Hs05049821_s1 (KRT83), and Hs99999905_m1 (GAPDH) (Thermo Fisher Scientific). All PCR reactions were performed in quadruplicate. Relative quantification of gene expression was performed using the 2−ΔΔCt method [40]. Fold change values were calculated using GAPDH as the internal control and an AA patient without T allele of rs142986308 was used as a calibrator.
2.23. Data and materials availability
The complete DNA microarray data set from expression analysis using mouse skins and patient hairs has been deposited in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/info/linking.html).
3. Results
3.1. Identification of AA susceptibility gene within the MHC region by haplotype sequencing
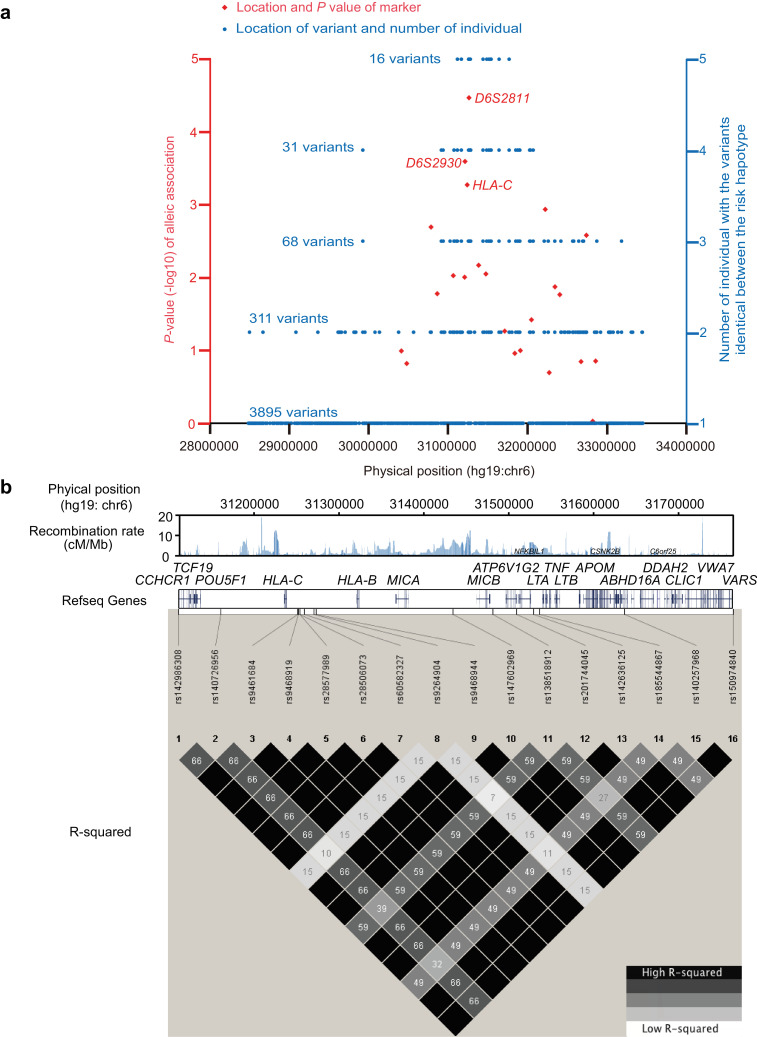
A total of 171 AA patients and 560 healthy controls were enrolled for association analysis using 22 microsatellites spanning the human leukocyte antigen (HLA) class I and II regions (chr6: 30,407,655- 32,854,116, hg19). We detected a single microsatellite, D6S2811 (allele 208, OR = 3.41, CI 95% = 1.94–5.99, P = 3.39 × 10−5), which was shown to be significantly associated with AA after Bonferroni correction (Fig. 1(a), Supplementary Table 4 and 5). Pair-wise evaluation of these multi-allelic loci indicated that a strong long-range LD was maintained across the assayed region of the MHC (Supplementary Fig. 5). Furthermore, estimation of haplotypes in 3 loci from D6S2811 to D6S2930 showed that the risk haplotype was defined as the segment that displayed a significant association with AA (MShap01, OR = 3.78, CI 95% = 2.00 - 7.16, P = 6.57 × 10−5) (Table 1 and Supplementary Table 6).
Fig. 1.
(Continued).
Fig. 1.
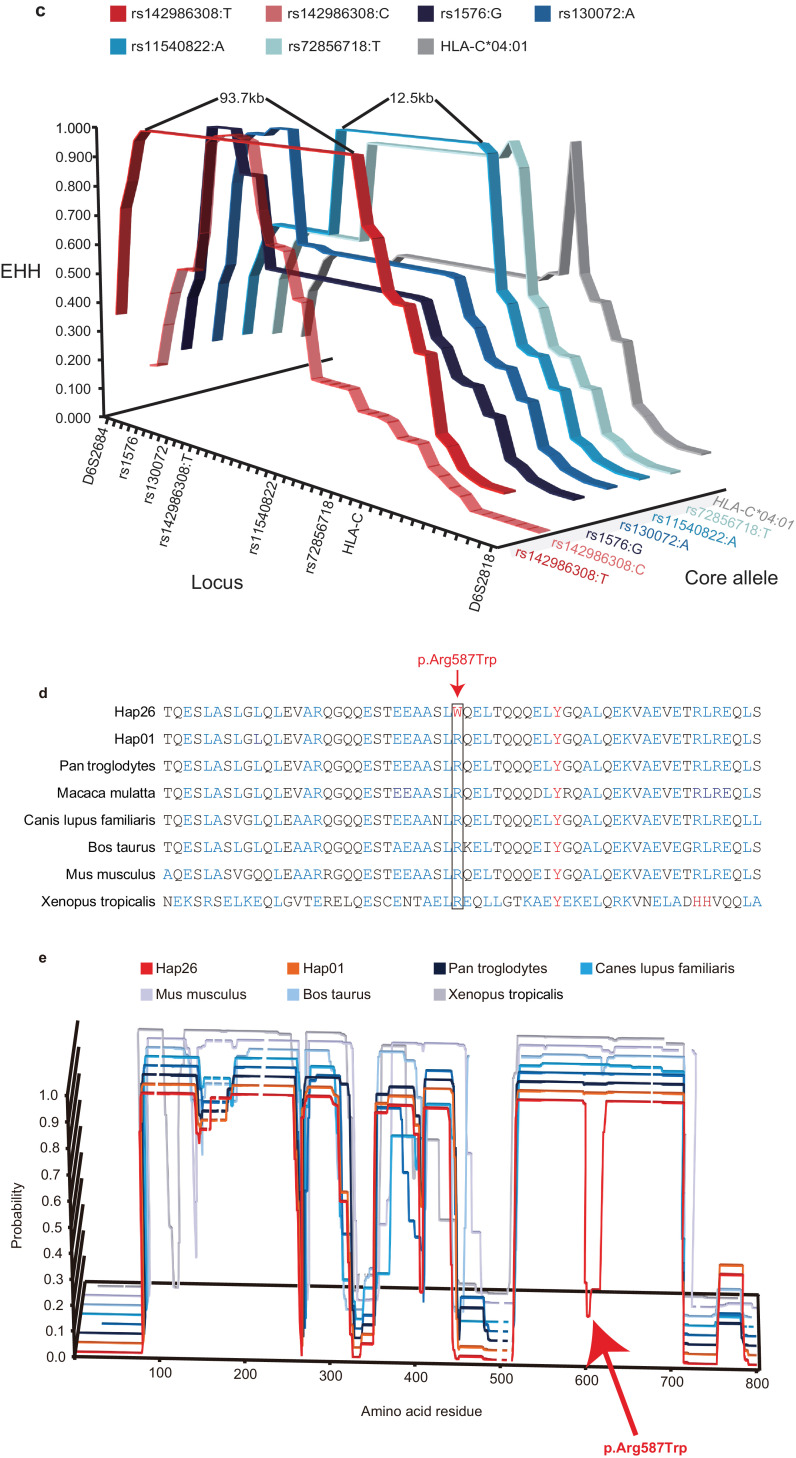
Identification of AA susceptibility gene within the MHC region by haplotype sequencing. (a) Association analysis and risk haplotype resequencing of the MHC region. Red diamonds indicate P values (-log10 scale) and locations. Three diamonds refer to haplotypes used for downstream analysis. Blue circles indicate individuals with variants identical between risk haplotype cases and the variant locations. (b) Pair-wise LD between 16 variants identified by NGS using genotype data from89 Japanese individuals obtained with the 1000 Genomes Browser. Upper track shows recombination rate (cM/Mb) estimated from Phase II HapMap data (release 21), middle track the gene map (RefSeq genes) generated with the UCSC Genome Browser, and lower track the pair-wise LD between the 16 variants in R-squared. (c) EHH analysis of core alleles at 43 loci displaying LD with the T allele of rs142986308. An estimated 43 loci haplotypes encompassing 24 SNVs of CCHCR1 and 19 multi-allelic loci (2.32Mbp) were used for this investigation. The 7 selected core alleles were as follows:rs142986308 allele T, rs142986308 allele C for the internal control, 4 SNVs that displayed LD with rs142986308 (Supplementary Fig. 28), and HLA-C*04:01 (Supplementary Fig. 3) as functional variants. (d) Multiple amino acid sequence alignment of CCHCR1 showing evolutionarily conserved amino acids. The sequences, except for Hap01 and Hap26, were NP_001009009 (Pan troglodytes), NP_001108422 (Macaca mulatta), XP_532,064(Canis lupus familiaris), NP_001019707 (Bos taurus), NP_666,360 (Mus musculus), and NP_001116918 (Xenopus tropicalis). Blue indicates residues that prefer to form coiled-coil domains (Ala, Glu, Lys, Leu, Arg) and red indicates aromatic residues that do not prefer to form coiled-coil domains. Arrow and box indicate the position of substitution of p.Arg587Trp (rs142986308). (e) Coiled-coil structure prediction of CCHCR1 in the AA-associated haplotype and different species using COILS v2.2. The Y axis indicates the probability of coiled-coil conformation and the X axis amino acid residue number. Full amino acid sequences are described in Supplementary notes. Multiple amino acid sequence alignments were assigned to the probabilities of coiled-coil conformation for each haplotype and specie. Line brakes correspond to gaps in the multiple alignments. Arrow shows position of p.Arg587Trp (rs142986308).
Table 1.
Haplotype association analysis of 3 loci around HLA-C gene.
| Haplotype | Haplotype frequency | Haplotype association | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype ID | D6S2930 | – | HLA-C | – | D6S2811 | Case | Control | OR | (95% CI) | P value |
| MShap01 | 441 | – | C*04:01 | – | 208 | 0.0614 | 0.0179 | 3.78 | 2.00 - 7.16 | 6.57 × 10−5 |
| MShap02 | 433 | – | C*07:02 | – | 192 | 0.0117 | 0.0009 | 13.4 | 1.49 - 121 | 7.24 × 10−3 |
| MShap03 | 437 | – | C*07:02 | – | 192 | 0.0292 | 0.0125 | 2.42 | 1.06 - 5.56 | 4.37 × 10−2 |
| MShap04 | 441 | – | C*15:02 | – | 192 | 0.0175 | 0.0018 | 10.2 | 2.03 - 50.7 | 1.91 × 10−3 |
Haplotypes in case subjects with a P-value <0.05 and haplotype frequency >0.01.
To move from the identified risk haplotypes to putative causal AA variants, we next sequenced 5 individuals with MShap01 and 7 individuals with the other non-risk haplotypes spanning the entire MHC (chr6:28,477,797–33,451,433, hg19) (Supplementary Table 2). As all risk haplotypes were heterozygous, any AA causal variant(s) would be expected to be heterozygous as well. Therefore, variants were filtered accordingly and only variants found to be identical between risk haplotypes were retained. Following this strategy, we extracted 3895 heterozygous risk variants from the 77,040 variants identified in the 12 individuals (Supplementary Table 6). Of these, only 16 variants were identical between the 5 AA risk haplotypes (Fig. 1(a), Table 2, Supplementary Fig. 2 and Supplementary Table 7) and only one was a non-synonymous coding SNV (rs142986308), defining a p.Arg587Trp substitution and mapping to CCHCR1 (Supplementary Fig. 6). Using pair-wise LD analysis, we further established that a haplotype composed of 16 extracted variants in a 651.7-kb region between CCHCR1 and VARS displayed strong LD despite including segments with a high recombination rate (Fig. 1(b)), implying that the haplotype was likely to be younger and/or has undergone positive selective pressure [16]. These results also suggest that our strategy used for stratification, sequencing, and filtering is effective for discovering risk haplotypes and novel MHC variants associated with AA.
Table 2.
Heterozygous variants identical in individuals with risk haplotype.
| Number of individual with risk haplotype | |||||
|---|---|---|---|---|---|
| Variant type | ≥ 1 | ≥ 2 | ≥ 3 | ≥ 4 | = 5 |
| Intergenic or intronic | 3736 | 298 | 65 | 29 | 15 |
| 5′ or 3′ UTR | 64 | 5 | 2 | 1 | 0 |
| Nonsense | 4 | 0 | 0 | 0 | 0 |
| Nonsynonymous | 51 | 5 | 1 | 1 | 1 |
| Synonymous | 40 | 3 | 0 | 0 | 0 |
| Total | 3895 | 311 | 68 | 31 | 16 |
To check for further variations and confirm the AA susceptibility variant in CCHCR1, we sequenced all coding exons in all subjects and found 22 variants, though only SNV rs142986308 was shared between all 5 patients and demonstrated a significant association with AA (OR = 3.41, CI 95% = 1.94–5.99, P = 3.39 × 10−5) (Supplementary Fig. 7 and Supplementary Table 8). Moreover, we estimated haplotypes for 24 SNVs. Haplotype 26 (Hap26) harboring the T allele rs142986308 showed a statistically significant association with AA (OR = 3.41, CI 95% = 1.94–5.99, P = 3.39 × 10−5), and rs142986308 was the only SNV associated with AA in Hap26 (Supplementary Table 9). Thus, we considered rs142986308 to be the primary variant associated with AA, though there were no observable correlations between the variant and disease severity.
Next, we investigated decay of the risk haplotype by recombination events that have occurred in evolutionary history. Extended haplotype homozygosity (EHH) analysis with 5 of the identified risk alleles (designated as core alleles in Fig. 1(c)) was conducted [41]. The core allele T of rs142986308 tagged the largest (93.7 kb) LD block with all values between rs1576 and D6S2931 showing EHH=1.00 Fig. 1(c)), indicating that Hap26, exclusively shared by the patients, had recently increased in frequency. Adjacent alleles also showed some frequency increase in the patient group, possibly by hitchhiking (Supplementary Table 8). Moreover, the variant rs142986308 was observed only in East Asian (allele frequency: 0.0277) (Supplementary Table 10), suggesting it to be population-specific. Although the exact selection mechanism operating on the T allele of rs142986308 remains unknown and the overall number of haplotypes analyzed is modest, it is plausible that this allele is the primary target of selection for AA. Hence, we excluded the other nonsense and non-synonymous variants as causal for AA, despite being in strong LD with the causal rs142986308 variant identified by haplotype and EHH analyses.
We next examined whether the variant rs142986308 had influence on the function of the CCHCR1 protein, which is predicted to contain several coiled-coil domains [42]. The domain including the AA-associated variant p.Arg587Trp is well conserved in parts across many species (Fig. 1(d) and Supplementary Table 11). Using structure prediction, the probability of coiled-coil conformation of this domain was notably reduced in only Hap26 harboring the AA-associated T allele of rs142986308 (Fig. 1(e), Supplementary Fig. 8 and 9). Aromatic substitutions are known to be more disruptive towards coiled-coil domains than alanine, glutamic acid, lysine, leucine, and arginine, which favor coiled-coil domain formation [43], adding weight to our speculation that variant rs142986308, which substitutes arginine with aromatic tryptophan, does indeed impair coiled-coil conformation of CCHCR1 (Fig. 1(d)).
We also performed homology searches using the PDB database and selected the best template structure for the partial CCHCR1 protein around the p.Arg587Trp substitution for homology modeling of the crystal structure of the human lamin-B1 coil 2 segment (Supplementary Fig. 10) [37]. A partial CCHCR1 structure was then generated using the template and utilized to evaluate the effect of p.Arg587Trp substitution on protein stability by performing molecular dynamics simulations with a CHARMm force field. The simulation findings showed that a p.Arg587Trp substitution reduced the stability of the protein (mutation energy: 1.03 kcal/mol) (Supplementary Fig. 11) and changed the CCHCR1 structure around the residue at 587 (Supplementary Fig. 12 and 13), indicating that this substitution may alter the protein-protein interaction of CCHCR1 (Supplementary Fig. 10).
3.2. Alopecic mice produced by allele-specific genome editing using CRISPR/Cas9
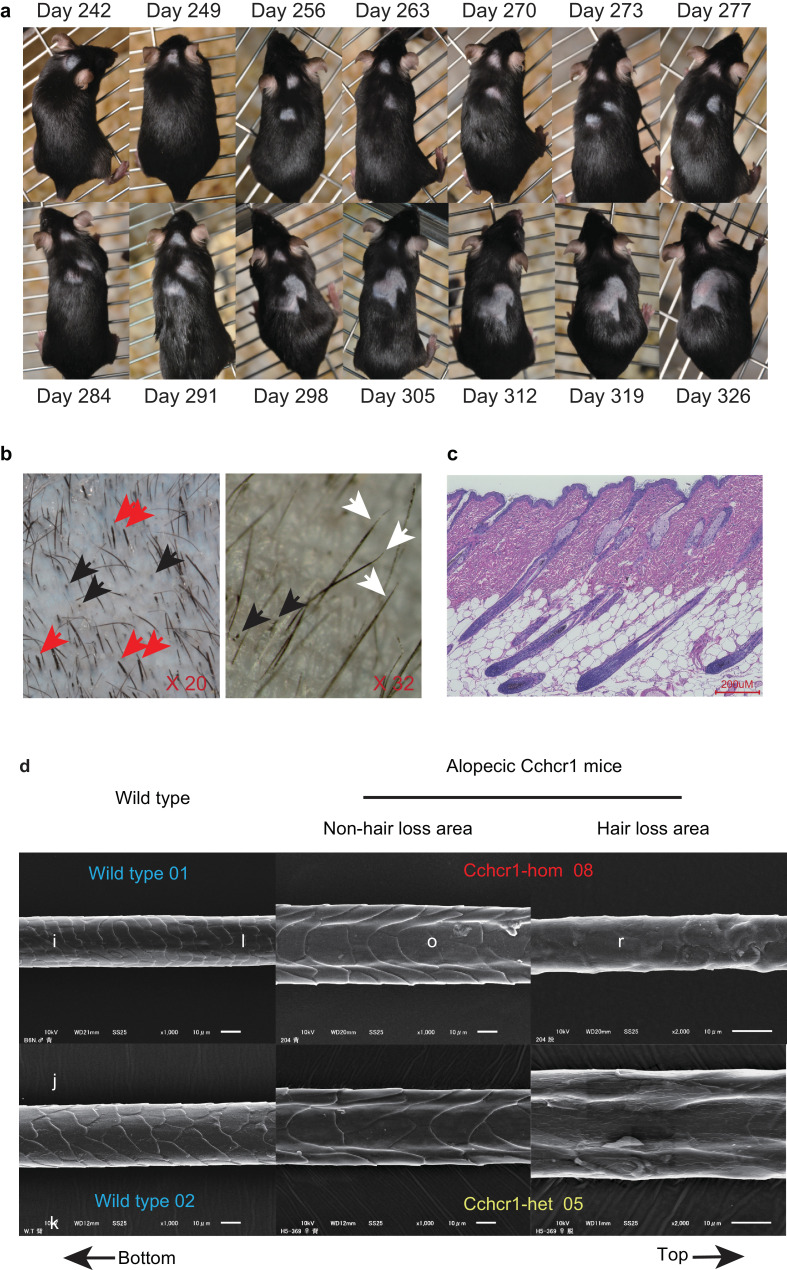
Next, we functionally evaluated the AA-associated variant by in vivo phenocopying p.Arg591Trp in murine Cchcr1 using allele-specific genome editing with the CRISPR/Cas9 system. Mice were generated with the risk allele concordant with p.Arg587Trp derived from the T allele of rs142986308 in humans (Supplementary Fig. 4), then we established mouse strains (Cchcr1 mice) with homozygous (Cchcr1-hom mice) and heterozygous risk alleles (Cchcr1-het mice). We also confirmed that this variant was not included among any of the mouse strains in the Sanger Mouse SNPs database (https://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1505). On the other hand, Cchcr1 knockout mice have been generated and listed [International Mouse Phenotyping Consortium (https://www.mousephenotype.org/), Mouse Genome Informatics (http://www.informatics.jax.org/)], though have yet to be phenotyped. In the present study, 2 (12.5%) of the 16 Cchcr1-het mice displayed hair loss, while 15 (55.5%) of the 27 Cchcr1-hom mice displayed hair loss patches after birth (assessed for up to 10 months), demonstrating that the risk allele induces hair loss. The incidence of hair loss was also higher in those mice as compared to C3H/HeJ mice, which showed spontaneous development of AA with age [44]. Over time, the initial area of hair loss expanded in the majority of the Cchcr1-mice (Fig. 2(a)), although constant and recovered hair loss was observed in some of those mice (Supplementary Fig. 14 and 15). Moreover, hair loss was not confined to certain areas, but was varied in the alopecic mice (Supplementary Fig. 16). The male to female ratio of alopecic Cchcr1 mice was nearly equal, and their surface displayed black spots, while the hair appeared to be broken and tapering (Fig. 2(b)), similar to the conditions seen in humans with AA and specific for AA-associated hair loss [45]. All of the Cchcr1 mice retained hair follicles in the area of hair loss and no signs of lymphocyte infiltration were seen in microscopic observations by HE staining and immunostaining using anti-CD4 and -CD8 antibodies (Fig. 2(c) and Supplementary Fig. 17), implying involvement of a non-immune mechanism in these cases. On the other hand, high-resolution scanning electron microscopy (SEM) identified hair abnormalities of cuticle formation in the alopecic Cchcr1-hom and -het mice. These mice displayed aberrant hair not only in areas of hair loss, but also in normal areas (Fig. 2(d)), suggesting that Cchcr1 mice are affected by abnormal hair keratinization.
Fig. 2.
Alopecic mice produced by allele-specific genome editing using CRISPR/Cas9. (a) Expansion of hair loss area in representative Cchcr1-hom mouse. (b) Morphology of hair loss area in Cchcr1-hom mouse. Red arrows show broken hair, black arrows show blackspots, white arrows show tapering hair. (c) Microscopic features of hair loss area in representative Cchcr1-hom mouse. Paraffin section of skin from representative Cchcr1-hom mouse after staining with hematoxylin and eosin. (d) Scanning electron microscopy (SEM) imaging of hair shafts was performed. In each panel, hair orientation is shown with the bottom to the left and top to the right.
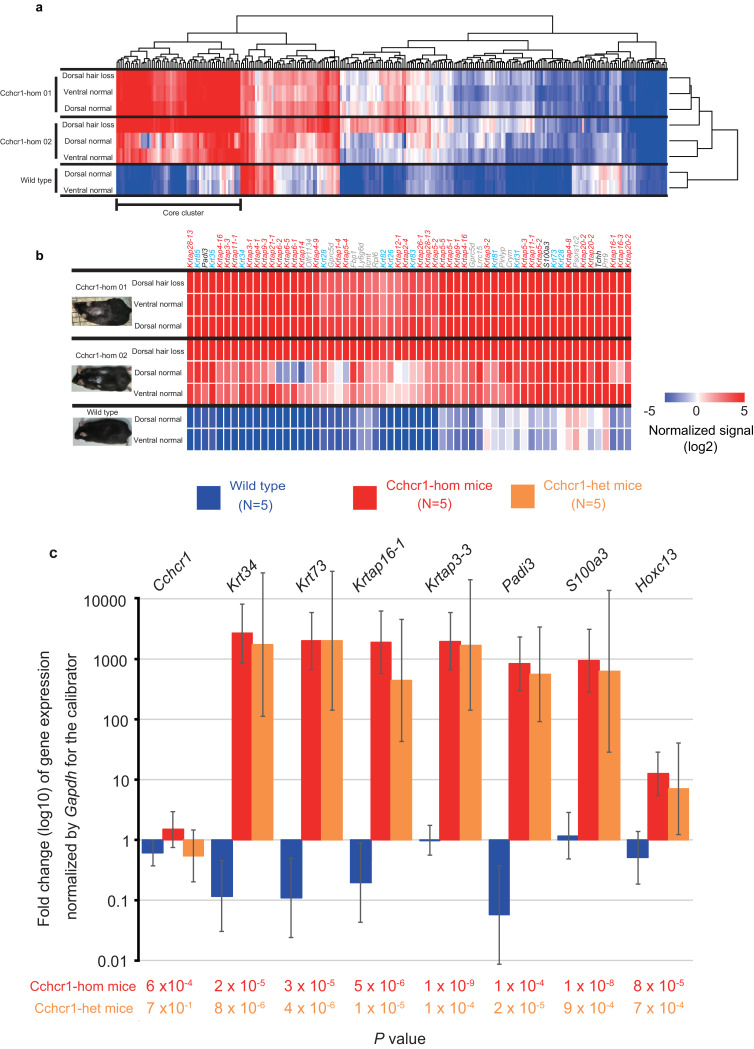
3.3. Expression analysis of alopecic Cchcr1 mice
To investigate a possible biological function underlying the observed hair loss in the Cchcr1 mice, we performed gene expression microarray analysis of dorsal and ventral skin biopsies of Cchcr1-hom mice (Supplementary Fig. 18 and 19), which identified 265 probes (246 genes) with 2-fold or greater up- or down-regulation as compared to wild-type mice (Supplementary Fig. 20 and Supplementary Table 12). Clustering analysis of these probes uncovered a strongly up-regulated gene cluster in the Cchcr1-hom mice (Fig. 3(a)), including hair-related genes (Fig. 3(b)). Nearly all of the up-regulated genes with greater 25-fold change were keratin (n = 12) and KRTAP (n = 31) genes (Supplementary Table 12). Hair keratins and KRTAPs are the major structural components of the hair shaft, and specifically expressed in the medulla, cortex, and cuticle layers of the shaft [46,47], Interaction between hair keratins and KRTAPs contributes to hair shaft rigidity [48].
Fig. 3.
(Continued).
Fig. 3.
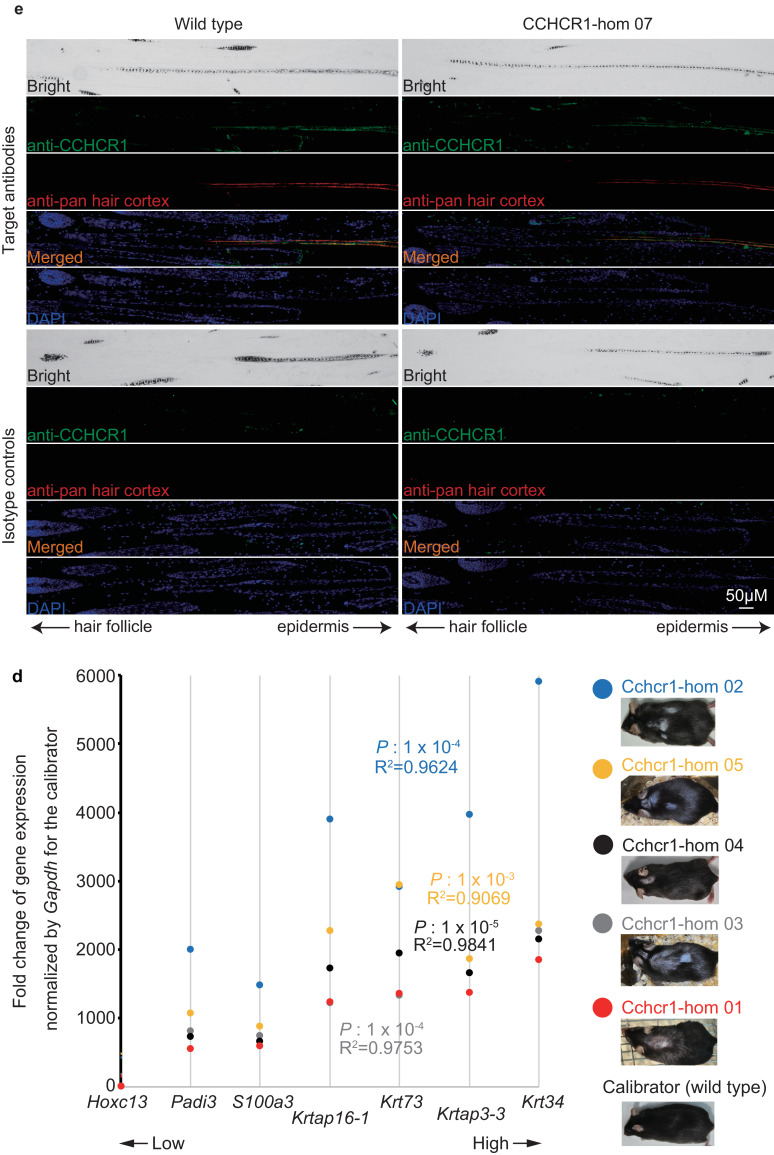
Expression analysis of Cchcr1 mice. (a) Heat map of 265 probes showing ≥ 2-fold change in gene expression. The list of genes is shown in Supplementary Table 12. The cluster displaying high expression in the dorsal hair loss area in both Cchcr1-hom mice was defined as the ‘core cluster’. (b) Heat map of core cluster genes. The color code depicts KRTAP family (red), keratin family (blue), other hair-related (black), and non hair-related (gray) genes. (c) Validation of upregulated gene expression for Cchcr1 and 7 selected genes. Mouse dorsal skin biopsies were subjected to expression analysis by qPCR and a comparative CT method. Bars reflect 95% confidence intervals. Fold change values were normalized to dorsal hair loss in a wild-type mouse as a calibrator, thus the fold change value of the calibrator was always one. Statistical significance was determined using Welch's t-test. (d) Gene expression trends in skin biopsies from dorsal hair loss areas in Cchcr1-hom mice. Correlation coefficients and the statistical significance was determined using Pearson's product-moment correlation between each mouse and Cchcr1-hom 01. (e) Co-localization of CCHCR1 with hair cortex keratin in follicles from Cchcr1-hom and wild-type mice skins. Paraffin sections were stained with anti-CCHCR1 (green) and anti-pan hair cortex keratin (red) antibodies, and subjected to fluorescent microscopy. Nuclear staining was performed by DAPI (Blue). Negative controls were stained with the isotype antibodies for the primary target antibodies. Each panel shows longitudinal section of skin including subcutaneous tissue along hair shafts. Hair orientation is shown with hair follicle to the left and epidermis to the right.
Other up-regulated genes included peptidyl arginine deaminase type III (Padi3), S100 calcium binding protein A3 (S100A3), trichohyalin (Tchh), and homeobox C13 (Hoxc13) (Supplementary Table 12) [49,50]. In cuticular cells, S100A3 is a substrate of PADI3, while in inner root sheath (IRS) cells of hair follicles it is a substrate of TCHH [49]. Ca2+-dependent modifications of S100A3 and TCHH by PADI3 play important roles in shaping and mechanically strengthening hair with keratin [49,51]. Hoxc13 is unique among the Hox genes, as it is expressed in the outer root sheath (ORS), matrix, medulla, and IRS of hair follicles in a hair cycle-dependent manner, and it has a role in hair shaft differentiation [50]. Thus, the majority of genes shown to be strongly up-regulated in Cchcr1-hom mice were involved in the hair shaft and its formation.
To confirm the microarray results, we performed quantitative PCR (qPCR) analysis of 7 of the up-regulated genes and Cchcr1 using a comparative CT method [40]. Those results confirmed significant differences between dorsal skin biopsies of the Cchcr1-hom and wild-type mice, and also indicated similar differences in Cchcr1-het mice (Fig. 3(c) and Supplementary Fig. 21).
All Cchcr1-hom mice showed highly concordant rank orders of expression levels of the examined genes (Fig. 3(d) and Supplementary Fig. 22–24), while Cchcr1-het mice showed moderate trends only. These results indicate involvement of regulatory networks in hair shaft differentiation[52].
Next, we evaluated the localization of CCHCR1 to delineate potential mechanisms underlying the observed hair loss in our mouse model. Immunostaining revealed CCHCR1 to be located in the mid-to-upper hair shaft but not in the hair shaft within the hair bulb (Fig. 3(e)), suggesting that CCHCR1 is a structural component of the hair shaft, similar to keratin. CCHCR1 was co-localized with the hair cortex and also found to be localized in the hair medulla (Fig. 3(e)), although no differences between Cchcr1-hom and wild type mice were observed. Furthermore, we demonstrated by Proximity Ligation Assay that the observed co-localization of CCHCR1 with hair cortex keratin was due to protein–protein interaction (Supplementary Fig. 25).
3.4. Functional analysis using hairs of AA patients with T allele of rs142986308
Finally, to validate the AA susceptibility variant not only by using mouse models but also human subjects, we evaluated hairs around hair loss areas of AA patients with the T allele of rs142986308 by SEM and gene expression microarray analysis.
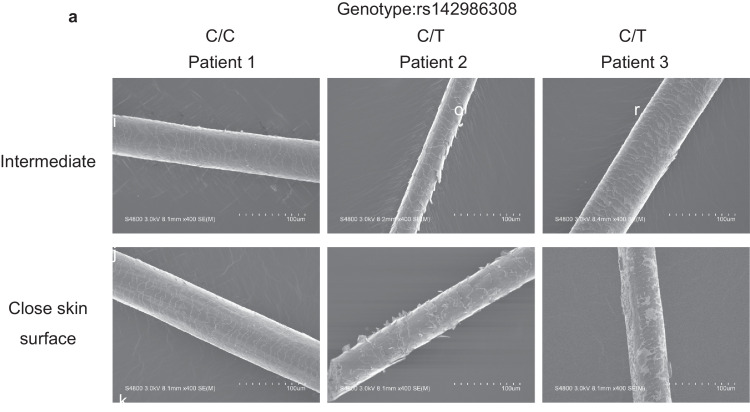
The SEM identified hair abnormalities of cuticle formation in AA patients with the T allele of rs142986308 (Fig. 4(a)) as well as the alopecic Cchcr1 mice (Fig. 2(d)), suggesting that AA patients with the T allele of rs142986308 are affected by abnormal hair keratinization. Previous studies also showed that the hair cuticles of AA patients are normal up to the point of breakage and suggested a transient malformation of the cortex [53,54].
Fig. 4.
(Continued).
Fig. 4.
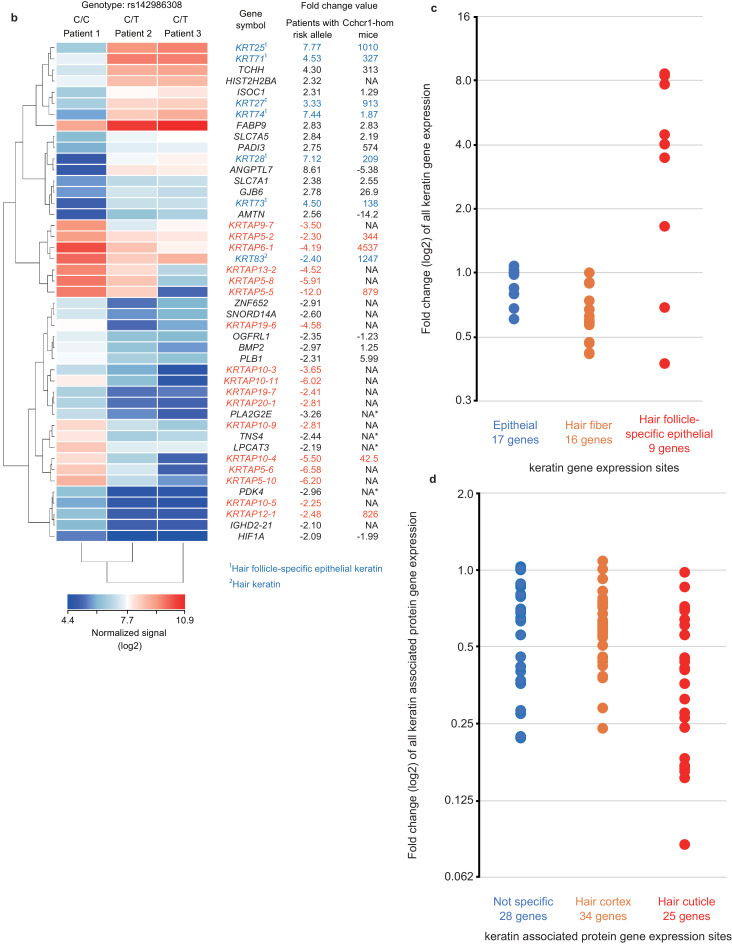
Functional analysis using hairs of AA patients with T allele of rs142986308. (a) Scanning electron microscopy (SEM) imaging of hair shafts was performed using AA patients with the T allele of rs142986308. (b) Microarray analysis using hair follicles of AA patients with T allele of rs142986308.Heat map of 45 genes showing ≥ 2-fold change in gene expression between the genotype ofrs142986308. Fold change value in the microarray analysis of Cchcr1-hom mice is also indicated. NA indicates missing value because of no probe concordant in the probe set of the mouse microarray platform in this study. NA* indicates missing value because the regulation of fold change values is not concordant between Cchcr1-hom mice. (c) Fold change values of all keratin gene expression in AA patients with rs142986308 C/T genotype against C/C genotype. Ten keratin genes are excluded because the regulation is inconsistent between AA patients. (d) Fold change values of all keratin associated protein gene expression in AA patients with rs142986308 C/T genotype against C/C genotype. Four keratin associated protein genes are excluded because the regulation is inconsistent between AA patients.
Gene expression microarray analysis using RNA from hair follicles identified 45 genes with 2-fold or greater up- or down-regulation as compared to an AA patient without the allele (Fig. 4(b), Supplementary Fig. 26, and Supplementary Table 13). These results were confirmed by qPCR analysis on 4 randomly selected genes out of the 45 genes (Supplementary Fig. 27). The majority of the 45 genes were keratin (n = 7) and KRTAP (n = 17) genes, indicating that these results were similar to the gene expression profiling of the Cchcr1 mice (Fig. 3(b) and 4(b)). PADI3 and TCHH were also included in them. However, all of the 17 KRTAP genes were down-regulated. Moreover, all up-regulated keratin genes were categorized as hair follicle-specific epithelial keratin and expressed in compartments of IRS (Fig. 4(c)) which plays an important role in the differentiation and keratinization of the matrix cells in the anagen hair bulb [55,56]. The down-regulated KRTAP genes were expressed in middle and upper keratogeneous zone of the hair fiber cuticle (Fig. 4(d)) [57]. Previous reports also indicated that these genes showed significantly differential expression between lesional scalps and control scalps in addition to many immune genes [58,59]. Thus, the functional analysis of Cchcr1 mice and AA patients implicated the variant we identified by genome analysis in aberrant keratinization and hair loss.
4. Discussion
Our finding that CCHCR1 is an AA-associated locus in the HLA class I region differs from that of a previous genome-wide association study, which suggested that HLA-DR. in the HLA class II region is a key driver of AA etiology [13]. One possible explanation for this seeming contradiction is that our study may not have been sufficiently powered to detect the weaker association of AA with the HLA-class II region, although the microsatellites used covered the entire class II region as well (Fig. 1(a)). On the other hand, consistent with our finding, studies performed in Chinese using HLA genes as markers suggest that the locus associated with AA maps to the class I rather than the class II region [60], [61], [62]. Moreover, variant rs142986308 is very rare in Caucasians (Supplementary Table 10) and has undergone positive selection (Fig. 1(c)), at least in Japanese, suggesting population-specific differences regarding AA risk haplotypes. This is the first study to show that an HLA class I allele, rs142986308, can be functionally linked to the hair loss phenotype, which has not been demonstrated for any other variants, including HLA-DR. …
Previous studies have provided evidence of a relationship between CCHCR1 and hair keratin-related genes. For example, a risk haplotype (CCHCR1*WWCC) was previously implicated to be involved in psoriasis in Europeans [42,63]. Transgenic mice with the risk haplotype appeared normal, although overexpression of CCHCR1 was shown to affect keratinocyte proliferation [64], and hyperproliferation of keratinocytes is a hallmark for psoriasis. However, keratin-related genes showed altered expression in mice at risk and most of the genes with significantly lower expression were those encoding hair keratins or KRTAPs [65]. These observations suggest that expression of these genes is dependent on the CCHCR1 haplotype.
C3H/HeJ mice have been recognized as a model that shows spontaneous development of adult onset AA [66]. This model demonstrates lymphocyte infiltration within hair follicles, follicular dystrophy, and spontaneous hair regrowth [66], though variants associated with the phenotypes have yet to be identified. On the other hand, a sub-strain of C57BL/6 different from that used in this study has also been reported to demonstrate hair loss and ulcerative dermatitis [67]. Nevertheless, we did not observe any evidence indicating inflammatory cell infiltration or dermatitis in Cchcr1 mice, thus the mechanism of hair loss in these strains may be different. To fully clarify the distinct pathogenic mechanisms of hair loss, more comprehensive characterization of these different mouse models is required.
PADI3 is known to be associated with central centrifugal cicatricial alopecia (CCCA) [68], autosomal recessive uncombable hair syndrome (UHS) [51], and shape variation of human head hair [69]. Missense mutations in PADI3, a causative gene of CCCA and UHS, induce formation of PADI3 protein aggregate with abnormal intracellular localization in the cytoplasm, and reduce expression and activity [51,68]. Moreover, findings of RNA-sequencing using scalp skin samples from CCCA patients as compared to control group samples revealed lower expressions not only in PADI3 but also in TCHH, many KRTAP genes, and KRT genes, all of which are involved in hair fiber development [68]. These findings are consistent with those obtained in the present study using Cchcr1 mice and AA patients with the risk rs142986308 variant, suggesting that this variant may also have an impact on the pathway of hair-shaft formation and contribute to hair loss.
Studies performed by Jackson et al. and Tobin et al. demonstrated that the hair cuticles of AA patients are normal up to the point of breakage [53,54], and suggested a transient malformation of the cortex. In contrast, the Cchcr1 mice used here showed aberrant hair cuticles as well as hair loss. Although we were not able to determine whether aberrant hair cuticles developed as a direct result of broken hairs in this study, any aberrant formation of the hair cortex may also contribute to hair loss, because CCHCR1 was observed in the cortex and the medulla. Moreover, we demonstrated that the variant identified by genetic analysis in AA patient genomes was related to a hair loss phenotype in Cchcr1 mouse skin, that CCHCR1 harboring the variant is a novel component of the hair shaft, and that the variant influenced the expression of genes involved in keratinization and development of hair in both mouse and human.
In summary, we clearly demonstrated that the variant identified by genetic analysis in AA patient genomes was related to a hair loss phenotype in Cchcr1 mouse skin, that CCHCR1 harboring the variant is a novel component of the hair shaft, and that the variant influenced the expression of genes involved in keratinization and development of hair in both mouse and human. Our findings point to an additional pathway for AA pathogenesis based on aberrant keratinization among many causative factors including immunological aspects, suggesting that the alopecia we observed in this study may be a subtype of AA generally defined as autoimmune disease. Moreover, our engineered mice provide a valuable resource for continued research into AA pathogenesis and the development of potential future treatments.
Acknowledgments
Acknowledgments
We express our gratefulness to Wang Ting, Hisako Kawada, and Hideki Hayashi of the Support Center for Medical Research and Education, Tokai University.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Founding Sources
This work was supported by JSPS KAKENHI (JP16K10177) and the NIHR UCLH Biomedical Research center (BRC84/CN/SB/5984). The funding sources had no role in the study design, data collection, data analysis, data interpretation, and writing of the report.
Author contributions
Akira Oka: conceived the studies, performed laboratory experiments, contributed to the data analysis, and drafted the manuscript. Atsushi Takagi, Etsuko Komiyama, Nagisa Yoshihara: were involved in sample collection and performed laboratory experiments. Shuhei Mano, Mahoko Takahashi Ueda, So Nakagawa, Masayuki Tanaka, Tomoyoshi Komiyama: were contributed to the data analysis and statistical support. Kazuyoshi Hosomichi, Shingo Suzuki, Yuko Haida, Nami Motosugi, Tomomi Hatanaka, Minoru Kimura, Hiromi Miura, Asako Otomo: performed laboratory experiments. Masato Ohtsuka: wrote the study protocols and contributed to supervision. Shinji Hadano, Tomotaka Mabuchi, Stephan Beck, Hidetoshi Inoko: were involved in reviewing and editing the manuscript. Shigaku Ikeda: conceived the studies, contributed to supervision and finalized the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102810.
Contributor Information
Akira Oka, Email: oka246@is.icc.u-tokai.ac.jp.
Shigaku Ikeda, Email: ikeda@juntendo.ac.jp.
Appendix. Supplementary materials
References
- 1.Wasserman D., Guzman Sanchez D., Scott K., McMichael A. Alopecia areata. International Journal of Dermatology. 2007;46(2):121–131. doi: 10.1111/j.1365-4632.2007.03193.x. [DOI] [PubMed] [Google Scholar]
- 2.Xing L., Dai Z., Jabbari A. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20(9):1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMichael A.J., Pearce D.J., Wasserman D. Alopecia in the United States: outpatient utilization and common prescribing patterns. J Am Acad Dermatol. 2007;57(2 Suppl):S49–S51. doi: 10.1016/j.jaad.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Jackow C., Puffer N., Hordinsky M., Nelson J., Tarrand J., Duvic M. Alopecia areata and cytomegalovirus infection in twins: Genes versus environment? Journal of the American Academy of Dermatology. 1998;38(3):418–425. doi: 10.1016/s0190-9622(98)70499-2. [DOI] [PubMed] [Google Scholar]
- 5.Yang S., Yang J., Liu J.B. The genetic epidemiology of alopecia areata in China. Br J Dermatol. 2004;151(1):16–23. doi: 10.1111/j.1365-2133.2004.05915.x. [DOI] [PubMed] [Google Scholar]
- 6.Tan E., Tay Y.-K., Goh C.-L., Chin Giam Y. The pattern and profile of alopecia areata in Singapore - a study of 219 Asians. International Journal of Dermatology. 2002;41(11):748–753. doi: 10.1046/j.1365-4362.2002.01357.x. [DOI] [PubMed] [Google Scholar]
- 7.van der Steen P., Traupe H., Happle R., Boezeman J., Strater R., Hamm H. The genetic risk for alopecia areata in first degree relatives of severely affected patients. An estimate. Acta Derm Venereol. 1992;72(5):373–375. [PubMed] [Google Scholar]
- 8.Shellow W.V., Edwards J.E., Koo J.Y. Profile of alopecia areata: a questionnaire analysis of patient and family. Int J Dermatol. 1992;31(3):186–189. doi: 10.1111/j.1365-4362.1992.tb03932.x. [DOI] [PubMed] [Google Scholar]
- 9.Xiao F.L., Yang S., Liu J.B. The epidemiology of childhood alopecia areata in China: a study of 226 patients. Pediatric dermatology. 2006;23(1):13–18. doi: 10.1111/j.1525-1470.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- 10.Muller C.S., El Shabrawi-Caelen L. 'Follicular Swiss cheese' pattern–another histopathologic clue to alopecia areata. J Cutan Pathol. 2011;38(2):185–189. doi: 10.1111/j.1600-0560.2010.01640.x. [DOI] [PubMed] [Google Scholar]
- 11.Phan K., Sebaratnam D.F. JAK inhibitors for alopecia areata: a systematic review and meta-analysis. Journal of the European Academy of Dermatology and Venereology: JEADV. 2019;33(5):850–856. doi: 10.1111/jdv.15489. [DOI] [PubMed] [Google Scholar]
- 12.Petukhova L., Duvic M., Hordinsky M. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betz R.C., Petukhova L., Ripke S. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. 2015;6:5966. doi: 10.1038/ncomms6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forstbauer L.M., Brockschmidt F.F., Moskvina V. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. European journal of human genetics: EJHG. 2012;20(3):326–332. doi: 10.1038/ejhg.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawashima M., Ohashi J., Nishida N., Tokunaga K. Evolutionary analysis of classical HLA class I and II genes suggests that recent positive selection acted on DPB1×04:01 in Japanese population. PLoS One. 2012;7(10):e46806. doi: 10.1371/journal.pone.0046806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bakker P.I., McVean G., Sabeti P.C. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulcher J. Microsatellite markers for linkage and association studies. Cold Spring Harb Protoc. 2012;2012(4):425–432. doi: 10.1101/pdb.top068510. [DOI] [PubMed] [Google Scholar]
- 18.Tamiya G., Shinya M., Imanishi T. Whole genome association study of rheumatoid arthritis using 27 039 microsatellites. Hum Mol Genet. 2005;14(16):2305–2321. doi: 10.1093/hmg/ddi234. [DOI] [PubMed] [Google Scholar]
- 19.Haida Y., Ikeda S., Takagi A. Association analysis of the HLA-C gene in Japanese alopecia areata. Immunogenetics. 2013;65(7):553–557. doi: 10.1007/s00251-013-0703-z. [DOI] [PubMed] [Google Scholar]
- 20.Cao H., Wu J., Wang Y. An integrated tool to study MHC region: accurate SNV detection and HLA genes typing in human MHC region using targeted high-throughput sequencing. PLoS One. 2013;8(7):e69388. doi: 10.1371/journal.pone.0069388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Handsaker B., Wysoker A. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A., Hanna M., Banks E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne I., Stephen G., Bayer M. Using Tablet for visual exploration of second-generation sequencing data. Briefings in bioinformatics. 2013;14(2):193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 26.de Cid R., Riveira-Munoz E., Zeeuwen P.L. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41(2):211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular ecology resources. 2008;8(1):103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 28.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Stephens M., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78(4):629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gautier M., Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 32.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 34.McDonnell A.V., Jiang T., Keating A.E., Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22(3):356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 35.Berger B., Wilson D.B., Wolf E., Tonchev T., Milla M., Kim P.S. Predicting coiled coils by use of pairwise residue correlations. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(18):8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S.F., Madden T.L., Schaffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan J., Xu C., Bian C. Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1. FEBS letters. 2012;586(4):314–318. doi: 10.1016/j.febslet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Harms D.W., Quadros R.M., Seruggia D. Mouse Genome Editing Using the CRISPR/Cas System. Curr Protoc Hum Genet. 2014;83 doi: 10.1002/0471142905.hg1507s83. 15 7 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura H., Gurumurthy C.B., Sato T., Sato M., Ohtsuka M. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Scientific reports. 2015;5 doi: 10.1038/srep12799. 12,799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Sabeti P.C., Reich D.E., Higgins J.M. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 42.Asumalahti K., Veal C., Laitinen T. Coding haplotype analysis supports HCR as the putative susceptibility gene for psoriasis at the MHC PSORS1 locus. Hum Mol Genet. 2002;11(5):589–597. doi: 10.1093/hmg/11.5.589. [DOI] [PubMed] [Google Scholar]
- 43.Gromiha M.M., Parry D.A. Characteristic features of amino acid residues in coiled-coil protein structures. Biophys Chem. 2004;111(2):95–103. doi: 10.1016/j.bpc.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 44.McElwee K.J., Hoffmann R. Alopecia areata - animal models. Clinical and experimental dermatology. 2002;27(5):410–417. doi: 10.1046/j.1365-2230.2002.01075.x. [DOI] [PubMed] [Google Scholar]
- 45.Inui S., Nakajima T., Nakagawa K., Itami S. Clinical significance of dermoscopy in alopecia areata: analysis of 300 cases. Int J Dermatol. 2008;47(7):688–693. doi: 10.1111/j.1365-4632.2008.03692.x. [DOI] [PubMed] [Google Scholar]
- 46.Fujikawa H., Fujimoto A., Farooq M., Ito M., Shimomura Y. Characterization of the human hair shaft cuticle-specific keratin-associated protein 10 family. J Invest Dermatol. 2013;133(12):2780–2782. doi: 10.1038/jid.2013.233. [DOI] [PubMed] [Google Scholar]
- 47.Langbein L., Rogers M.A., Praetzel-Wunder S., Helmke B., Schirmacher P., Schweizer J. K25 (K25irs1), K26 (K25irs2), K27 (K25irs3), and K28 (K25irs4) represent the type I inner root sheath keratins of the human hair follicle. J Invest Dermatol. 2006;126(11):2377–2386. doi: 10.1038/sj.jid.5700494. [DOI] [PubMed] [Google Scholar]
- 48.Fujikawa H., Fujimoto A., Farooq M., Ito M., Shimomura Y. Characterization of the human hair keratin-associated protein 2 (KRTAP2) gene family. J Invest Dermatol. 2012;132(7):1806–1813. doi: 10.1038/jid.2012.73. [DOI] [PubMed] [Google Scholar]
- 49.Kizawa K., Takahara H., Unno M., Heizmann C.W. S100 and S100 fused-type protein families in epidermal maturation with special focus on S100A3 in mammalian hair cuticles. Biochimie. 2011;93(12):2038–2047. doi: 10.1016/j.biochi.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 50.Qiu W., Lei M., Tang H. Hoxc13 is a crucial regulator of murine hair cycle. Cell Tissue Res. 2016;364(1):149–158. doi: 10.1007/s00441-015-2312-7. [DOI] [PubMed] [Google Scholar]
- 51.FB U.B., Cau L., Tafazzoli A. Mutations in Three Genes Encoding Proteins Involved in Hair Shaft Formation Cause Uncombable Hair Syndrome. Am J Hum Genet. 2016;99(6):1292–1304. doi: 10.1016/j.ajhg.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potter C.S., Pruett N.D., Kern M.J. The nude mutant gene Foxn1 is a HOXC13 regulatory target during hair follicle and nail differentiation. J Invest Dermatol. 2011;131(4):828–837. doi: 10.1038/jid.2010.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson D., Church R.E., Ebling F.J. Alopecia areata hairs: a scanning electron microscope study. Br J Dermatol. 1971;85(3):242–246. doi: 10.1111/j.1365-2133.1971.tb07224.x. [DOI] [PubMed] [Google Scholar]
- 54.Tobin D.J., Fenton D.A., Kendall M.D. Ultrastructural study of exclamation-mark hair shafts in alopecia areata. J Cutan Pathol. 1990;17(6):348–354. doi: 10.1111/j.1600-0560.1990.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 55.Schweizer J., Langbein L., Rogers M.A., Winter H. Hair follicle-specific keratins and their diseases. Exp Cell Res. 2007;313(10):2010–2020. doi: 10.1016/j.yexcr.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 56.Harel S., Christiano A.M. Genetics of Structural Hair Disorders. Journal of Investigative Dermatology. 2012;132:E22–EE6. doi: 10.1038/skinbio.2012.7. [DOI] [PubMed] [Google Scholar]
- 57.Shimomura Y., Ito M. Human hair keratin-associated proteins. J Investig Dermatol Symp Proc. 2005;10(3):230–233. doi: 10.1111/j.1087-0024.2005.10112.x. [DOI] [PubMed] [Google Scholar]
- 58.Suarez-Farinas M., Ungar B., Noda S. Alopecia areata profiling shows TH1, TH2, and IL-23 cytokine activation without parallel TH17/TH22 skewing. J Allergy Clin Immunol. 2015;136(5):1277–1287. doi: 10.1016/j.jaci.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 59.Subramanya R.D., Coda A.B., Sinha A.A. Transcriptional profiling in alopecia areata defines immune and cell cycle control related genes within disease-specific signatures. Genomics. 2010;96(3):146–153. doi: 10.1016/j.ygeno.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Xiao F.L., Zhou F.S., Liu J.B. Association of HLA-DQA1 and DQB1 alleles with alolpecia areata in Chinese Hans. Arch Dermatol Res. 2005;297(5):201–209. doi: 10.1007/s00403-005-0608-2. [DOI] [PubMed] [Google Scholar]
- 61.Xiao F.L., Yang S., Yan K.L. Association of HLA class I alleles with aloplecia areata in Chinese Hans. J Dermatol Sci. 2006;41(2):109–119. doi: 10.1016/j.jdermsci.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Xiao F.L., Yang S., Lin G.S. HLA haplotypic association with different phenotype of alopecia areata in Chinese Hans. J Dermatol Sci. 2007;45(3):206–209. doi: 10.1016/j.jdermsci.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Asumalahti K., Laitinen T., Itkonen-Vatjus R. A candidate gene for psoriasis near HLA-C, HCR (Pg8), is highly polymorphic with a disease-associated susceptibility allele. Hum Mol Genet. 2000;9(10):1533–1542. doi: 10.1093/hmg/9.10.1533. [DOI] [PubMed] [Google Scholar]
- 64.Tiala I., Wakkinen J., Suomela S. The PSORS1 locus gene CCHCR1 affects keratinocyte proliferation in transgenic mice. Hum Mol Genet. 2008;17(7):1043–1051. doi: 10.1093/hmg/ddm377. [DOI] [PubMed] [Google Scholar]
- 65.Elomaa O., Majuri I., Suomela S. Transgenic mouse models support HCR as an effector gene in the PSORS1 locus. Hum Mol Genet. 2004;13(15):1551–1561. doi: 10.1093/hmg/ddh178. [DOI] [PubMed] [Google Scholar]
- 66.Sun J., Silva K.A., McElwee K.J., King L.E., Jr., Sundberg J.P. The C3H/HeJ mouse and DEBR rat models for alopecia areata: review of preclinical drug screening approaches and results. Exp Dermatol. 2008;17(10):793–805. doi: 10.1111/j.1600-0625.2008.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sundberg J.P., Taylor D., Lorch G. Primary follicular dystrophy with scarring dermatitis in C57BL/6 mouse substrains resembles central centrifugal cicatricial alopecia in humans. Veterinary pathology. 2011;48(2):513–524. doi: 10.1177/0300985810379431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malki L., Sarig O., Romano M.T. Variant PADI3 in Central Centrifugal Cicatricial Alopecia. The New England journal of medicine. 2019;380(9):833–841. doi: 10.1056/NEJMoa1816614. [DOI] [PubMed] [Google Scholar]
- 69.Liu F., Chen Y., Zhu G. Meta-analysis of genome-wide association studies identifies 8 novel loci involved in shape variation of human head hair. Hum Mol Genet. 2018;27(3):559–575. doi: 10.1093/hmg/ddx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.