Fig. 4.
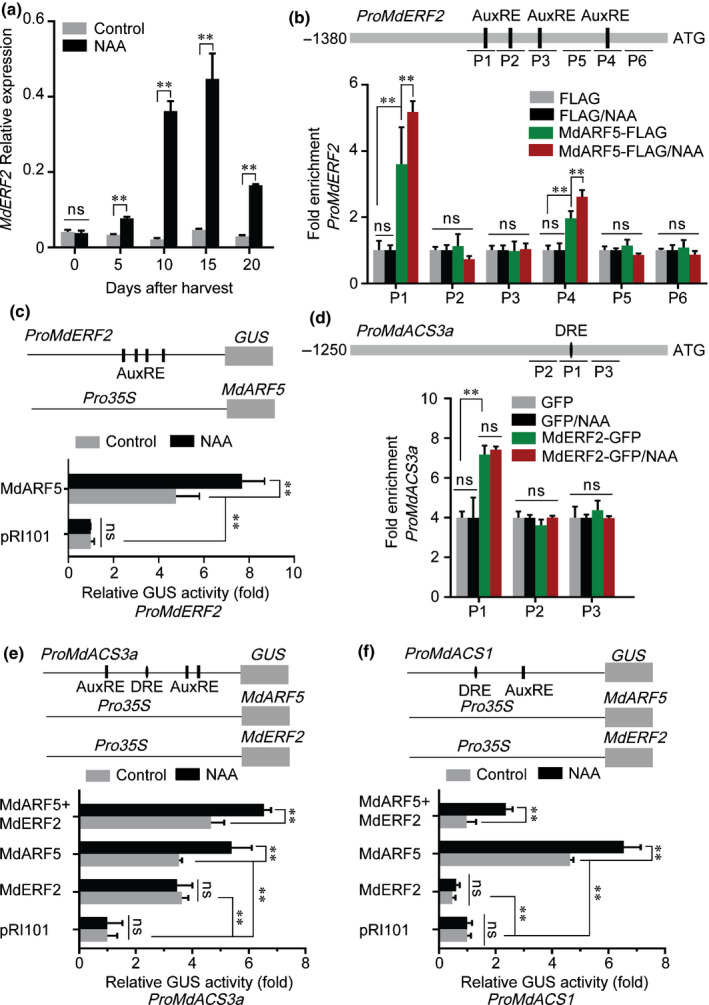
MdARF5 together with MdERF2 regulate ethylene biosynthesis. (a) The expression level of MdERF2 was determined by quantitative reverse transcription (qRT)‐PCR in apples treated with or without naphthaleneacetic acid (NAA). Fruit tissues were the same as in Fig. 1(f). Control, untreated fruit; NAA, NAA‐treated fruit. Three biological experiments with independently extracted RNA for each group of fruit were analyzed. Values represent means ± SE. Asterisks indicate significant differences as determined by a Student’s t‐test (**, P < 0.01); ns, no significant difference. (b) ChIP (chromatin immunoprecipitation)‐PCR assay showing MdARF5 binding to the MdERF2 promoter (1380 bp) in vivo. ChIP‐PCR was conducted as in Fig. 3(a). Six fragments (P1–P6) were analyzed. (c) GUS (β‐glucosidase) activation assay showing that MdARF5 positively regulates the MdERF2 promoter. The MdARF5 effector vector and MdERF2 promoter reporter vector were co‐infiltrated into wild tobacco leaves to analyze GUS activity. (d) ChIP‐PCR assay showing MdERF2 binding to the MdACS3a promoter (1250 bp) in vivo. Cross‐linked chromatin samples were extracted from MdERF2‐GFP (green fluorescent protein)‐overexpressing fruit calli, which were treated with or without NAA and precipitated with GFP antibody. Eluted DNA was used to amplify sequence neighboring the DRE‐motif (dehydration responsive element, ERF binding site) by qPCR. Three fragments (P1–P3) were analyzed. Fruit calli overexpressing the GFP sequence were used as a negative control. (e) GUS activation assay showing that MdARF5 together with MdERF2 promotes MdACS3a promoter activity. The MdARF5 and MdERF2 effector vectors were co‐infiltrated into wild tobacco leaves with the MdACS3a promoter effector vector, before analysis of GUS activity. (f) GUS activation assay showing that MdERF2 reduces MdACS1 promoter activity promoted by MdARF5. The MdARF5 and MdERF2 effector vectors were co‐infiltrated into wild tobacco leaves with the MdACS1 promoter effector vector to analyze GUS activity. For ChIP‐PCR, the ChIP assay was repeated three times and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. For the GUS activation assay, three independent transfections were analyzed. Values represent means ± SE. Asterisks indicate significant differences as determined by a Student’s t‐test (**, P < 0.01); ns, no significant difference.
